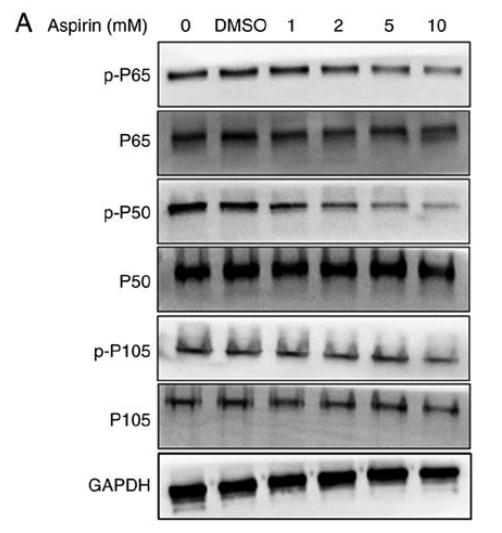
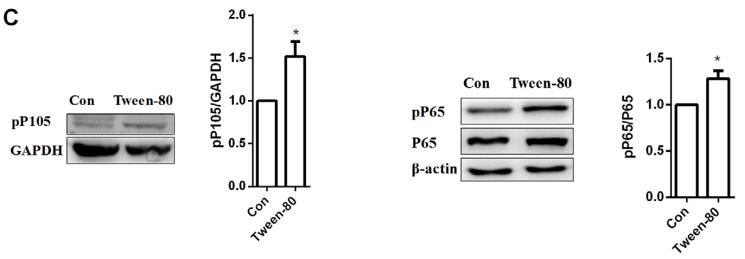
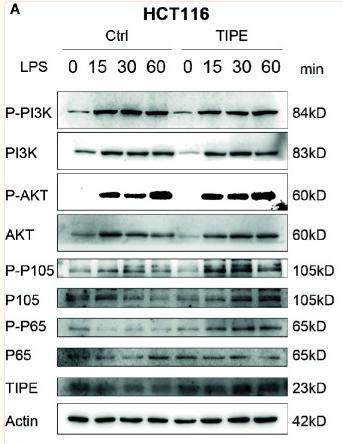
Phospho-NF kappaB p105/p50 (Ser337) Antibody - #AF3219
| Product: | Phospho-NF kappaB p105/p50 (Ser337) Antibody |
| Catalog: | AF3219 |
| Description: | Rabbit polyclonal antibody to Phospho-NF kappaB p105/p50 (Ser337) |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 105,50kDa; 105kD(Calculated). |
| Uniprot: | P19838 |
| RRID: | AB_2834647 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF3219, RRID:AB_2834647.
Fold/Unfold
DKFZp686C01211; DNA binding factor KBF1; DNA binding factor KBF1 EBP1; DNA-binding factor KBF1; EBP 1; EBP-1; EBP1; KBF1; MGC54151; NF kappa B; NF kappaB; NF kappabeta; NF kB1; NFkappaB; NFKB 1; NFKB p105; NFKB p50; Nfkb1; NFKB1_HUMAN; Nuclear factor kappa B DNA binding subunit; Nuclear factor kappa-B, subunit 1; Nuclear factor NF kappa B p105 subunit; Nuclear factor NF kappa B p50 subunit; Nuclear factor NF-kappa-B p50 subunit; Nuclear factor of kappa light chain gene enhancer in B cells 1; Nuclear factor of kappa light polypeptide gene enhancer in B cells 1; Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1; p105; p50; p84/NF-kappa-B1 p98; Transcription factor NFKB1;
Immunogens
A synthesized peptide derived from human NF- kappaB p105/p50 around the phosphorylation site of Ser337.
- P19838 NFKB1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAEDDPYLGRPEQMFHLDPSLTHTIFNPEVFQPQMALPTDGPYLQILEQPKQRGFRFRYVCEGPSHGGLPGASSEKNKKSYPQVKICNYVGPAKVIVQLVTNGKNIHLHAHSLVGKHCEDGICTVTAGPKDMVVGFANLGILHVTKKKVFETLEARMTEACIRGYNPGLLVHPDLAYLQAEGGGDRQLGDREKELIRQAALQQTKEMDLSVVRLMFTAFLPDSTGSFTRRLEPVVSDAIYDSKAPNASNLKIVRMDRTAGCVTGGEEIYLLCDKVQKDDIQIRFYEEEENGGVWEGFGDFSPTDVHRQFAIVFKTPKYKDINITKPASVFVQLRRKSDLETSEPKPFLYYPEIKDKEEVQRKRQKLMPNFSDSFGGGSGAGAGGGGMFGSGGGGGGTGSTGPGYSFPHYGFPTYGGITFHPGTTKSNAGMKHGTMDTESKKDPEGCDKSDDKNTVNLFGKVIETTEQDQEPSEATVGNGEVTLTYATGTKEESAGVQDNLFLEKAMQLAKRHANALFDYAVTGDVKMLLAVQRHLTAVQDENGDSVLHLAIIHLHSQLVRDLLEVTSGLISDDIINMRNDLYQTPLHLAVITKQEDVVEDLLRAGADLSLLDRLGNSVLHLAAKEGHDKVLSILLKHKKAALLLDHPNGDGLNAIHLAMMSNSLPCLLLLVAAGADVNAQEQKSGRTALHLAVEHDNISLAGCLLLEGDAHVDSTTYDGTTPLHIAAGRGSTRLAALLKAAGADPLVENFEPLYDLDDSWENAGEDEGVVPGTTPLDMATSWQVFDILNGKPYEPEFTSDDLLAQGDMKQLAEDVKLQLYKLLEIPDPDKNWATLAQKLGLGILNNAFRLSPAPSKTLMDNYEVSGGTVRELVEALRQMGYTEAIEVIQAASSPVKTTSQAHSLPLSPASTRQQIDELRDSDSVCDSGVETSFRKLSFTESLTSGASLLTLNKMPHDYGQEGPLEGKI
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
NF-kappa-B is a pleiotropic transcription factor present in almost all cell types and is the endpoint of a series of signal transduction events that are initiated by a vast array of stimuli related to many biological processes such as inflammation, immunity, differentiation, cell growth, tumorigenesis and apoptosis. NF-kappa-B is a homo- or heterodimeric complex formed by the Rel-like domain-containing proteins RELA/p65, RELB, NFKB1/p105, NFKB1/p50, REL and NFKB2/p52 and the heterodimeric p65-p50 complex appears to be most abundant one. The dimers bind at kappa-B sites in the DNA of their target genes and the individual dimers have distinct preferences for different kappa-B sites that they can bind with distinguishable affinity and specificity. Different dimer combinations act as transcriptional activators or repressors, respectively. NF-kappa-B is controlled by various mechanisms of post-translational modification and subcellular compartmentalization as well as by interactions with other cofactors or corepressors. NF-kappa-B complexes are held in the cytoplasm in an inactive state complexed with members of the NF-kappa-B inhibitor (I-kappa-B) family. In a conventional activation pathway, I-kappa-B is phosphorylated by I-kappa-B kinases (IKKs) in response to different activators, subsequently degraded thus liberating the active NF-kappa-B complex which translocates to the nucleus. NF-kappa-B heterodimeric p65-p50 and RelB-p50 complexes are transcriptional activators. The NF-kappa-B p50-p50 homodimer is a transcriptional repressor, but can act as a transcriptional activator when associated with BCL3. NFKB1 appears to have dual functions such as cytoplasmic retention of attached NF-kappa-B proteins by p105 and generation of p50 by a cotranslational processing. The proteasome-mediated process ensures the production of both p50 and p105 and preserves their independent function, although processing of NFKB1/p105 also appears to occur post-translationally. p50 binds to the kappa-B consensus sequence 5'-GGRNNYYCC-3', located in the enhancer region of genes involved in immune response and acute phase reactions. In a complex with MAP3K8, NFKB1/p105 represses MAP3K8-induced MAPK signaling; active MAP3K8 is released by proteasome-dependent degradation of NFKB1/p105.
While translation occurs, the particular unfolded structure after the GRR repeat promotes the generation of p50 making it an acceptable substrate for the proteasome. This process is known as cotranslational processing. The processed form is active and the unprocessed form acts as an inhibitor (I kappa B-like), being able to form cytosolic complexes with NF-kappa B, trapping it in the cytoplasm. Complete folding of the region downstream of the GRR repeat precludes processing.
Phosphorylation at 'Ser-903' and 'Ser-907' primes p105 for proteolytic processing in response to TNF-alpha stimulation. Phosphorylation at 'Ser-927' and 'Ser-932' are required for BTRC/BTRCP-mediated proteolysis.
Polyubiquitination seems to allow p105 processing.
S-nitrosylation of Cys-61 affects DNA binding.
The covalent modification of cysteine by 15-deoxy-Delta12,14-prostaglandin-J2 is autocatalytic and reversible. It may occur as an alternative to other cysteine modifications, such as S-nitrosylation and S-palmitoylation.
Nucleus. Cytoplasm.
Note: Nuclear, but also found in the cytoplasm in an inactive form complexed to an inhibitor (I-kappa-B).
The C-terminus of p105 might be involved in cytoplasmic retention, inhibition of DNA-binding, and transcription activation.
Glycine-rich region (GRR) appears to be a critical element in the generation of p50.
Research Fields
· Cellular Processes > Cell growth and death > Apoptosis. (View pathway)
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Ras signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cAMP signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > NF-kappa B signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > HIF-1 signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Sphingolipid signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > TNF signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > Antifolate resistance.
· Human Diseases > Endocrine and metabolic diseases > Insulin resistance.
· Human Diseases > Endocrine and metabolic diseases > Non-alcoholic fatty liver disease (NAFLD).
· Human Diseases > Substance dependence > Cocaine addiction.
· Human Diseases > Infectious diseases: Bacterial > Epithelial cell signaling in Helicobacter pylori infection.
· Human Diseases > Infectious diseases: Bacterial > Shigellosis.
· Human Diseases > Infectious diseases: Bacterial > Salmonella infection.
· Human Diseases > Infectious diseases: Bacterial > Pertussis.
· Human Diseases > Infectious diseases: Bacterial > Legionellosis.
· Human Diseases > Infectious diseases: Parasitic > Leishmaniasis.
· Human Diseases > Infectious diseases: Parasitic > Chagas disease (American trypanosomiasis).
· Human Diseases > Infectious diseases: Parasitic > Toxoplasmosis.
· Human Diseases > Infectious diseases: Parasitic > Amoebiasis.
· Human Diseases > Infectious diseases: Bacterial > Tuberculosis.
· Human Diseases > Infectious diseases: Viral > Hepatitis C.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Measles.
· Human Diseases > Infectious diseases: Viral > Influenza A.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Infectious diseases: Viral > Herpes simplex infection.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Pancreatic cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Prostate cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Chronic myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Acute myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Small cell lung cancer. (View pathway)
· Human Diseases > Immune diseases > Inflammatory bowel disease (IBD).
· Organismal Systems > Immune system > Chemokine signaling pathway. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway. (View pathway)
· Organismal Systems > Development > Osteoclast differentiation. (View pathway)
· Organismal Systems > Immune system > Toll-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > RIG-I-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Cytosolic DNA-sensing pathway. (View pathway)
· Organismal Systems > Immune system > IL-17 signaling pathway. (View pathway)
· Organismal Systems > Immune system > Th1 and Th2 cell differentiation. (View pathway)
· Organismal Systems > Immune system > Th17 cell differentiation. (View pathway)
· Organismal Systems > Immune system > T cell receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > B cell receptor signaling pathway. (View pathway)
· Organismal Systems > Nervous system > Neurotrophin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Prolactin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Adipocytokine signaling pathway.
· Organismal Systems > Endocrine system > Relaxin signaling pathway.
References
Application: WB Species: human Sample: RA‑FLS cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.