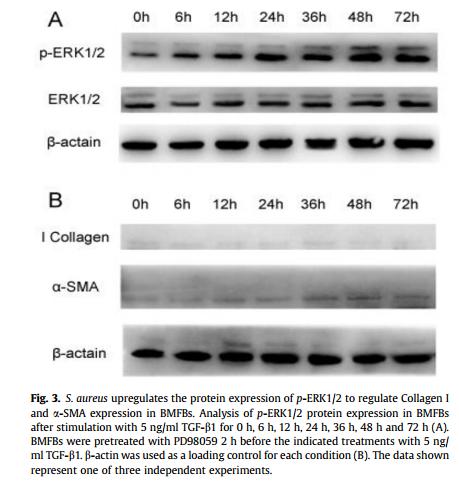
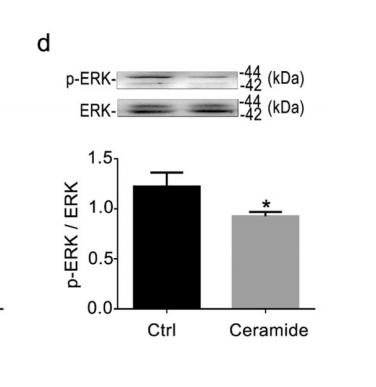
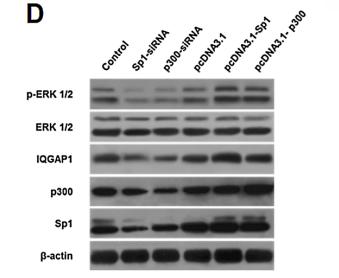
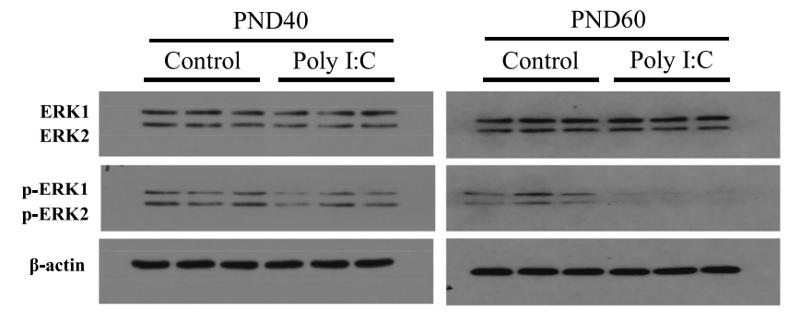
Phospho-ERK1/2 (Tyr204) Antibody - #AF1014
| Product: | Phospho-ERK1/2 (Tyr204) Antibody |
| Catalog: | AF1014 |
| Description: | Rabbit polyclonal antibody to Phospho-ERK1/2 (Tyr204) |
| Application: | WB IHC |
| Cited expt.: | WB, IHC |
| Reactivity: | Human, Mouse, Rat, Bovine |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit |
| Mol.Wt.: | 42,44kDa; 43kD,41kD(Calculated). |
| Uniprot: | P27361 | P28482 |
| RRID: | AB_2834412 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF1014, RRID:AB_2834412.
Fold/Unfold
ERK 1; ERK; ERK-1; ERK1; ERT 2; ERT2; Extracellular Signal Regulated Kinase 1; Extracellular signal related kinase 1; Extracellular signal-regulated kinase 1; HGNC6877; HS44KDAP; HUMKER1A; Insulin Stimulated MAP2 Kinase; Insulin-stimulated MAP2 kinase; MAP kinase 1; MAP kinase 3; MAP Kinase; MAP kinase isoform p44; MAPK 1; MAPK 3; MAPK; MAPK1; Mapk3; MGC20180; Microtubule Associated Protein 2 Kinase; Microtubule-associated protein 2 kinase; Mitogen Activated Protein Kinase 3; Mitogen-activated protein kinase 1; Mitogen-activated protein kinase 3; MK03_HUMAN; OTTHUMP00000174538; OTTHUMP00000174541; p44 ERK1; p44 MAPK; p44-ERK1; p44-MAPK; P44ERK1; P44MAPK; PRKM 3; PRKM3; Protein Kinase Mitogen Activated 3; ERK 2; ERK; ERK-2; ERT1; Extracellular Signal Regulated Kinase 2; Extracellular signal-regulated kinase 2; MAP kinase 1; MAP kinase 2; MAP kinase isoform p42; MAPK 1; MAPK 2; Mapk1; MAPK2; Mitogen-activated protein kinase 1; Mitogen-activated protein kinase 2; MK01_HUMAN; P38; P40; P41; p42-MAPK; P42MAPK; PRKM1; PRKM2; protein kinase, mitogen-activated, 1; protein kinase, mitogen-activated, 2; protein tyrosine kinase ERK2;
Immunogens
A synthesized peptide derived from human ERK1/2 around the phosphorylation site of Tyr204.
- P27361 MK03_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAAAAAQGGGGGEPRRTEGVGPGVPGEVEMVKGQPFDVGPRYTQLQYIGEGAYGMVSSAYDHVRKTRVAIKKISPFEHQTYCQRTLREIQILLRFRHENVIGIRDILRASTLEAMRDVYIVQDLMETDLYKLLKSQQLSNDHICYFLYQILRGLKYIHSANVLHRDLKPSNLLINTTCDLKICDFGLARIADPEHDHTGFLTEYVATRWYRAPEIMLNSKGYTKSIDIWSVGCILAEMLSNRPIFPGKHYLDQLNHILGILGSPSQEDLNCIINMKARNYLQSLPSKTKVAWAKLFPKSDSKALDLLDRMLTFNPNKRITVEEALAHPYLEQYYDPTDEPVAEEPFTFAMELDDLPKERLKELIFQETARFQPGVLEAP
- P28482 MK01_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAAAAAAGAGPEMVRGQVFDVGPRYTNLSYIGEGAYGMVCSAYDNVNKVRVAIKKISPFEHQTYCQRTLREIKILLRFRHENIIGINDIIRAPTIEQMKDVYIVQDLMETDLYKLLKTQHLSNDHICYFLYQILRGLKYIHSANVLHRDLKPSNLLLNTTCDLKICDFGLARVADPDHDHTGFLTEYVATRWYRAPEIMLNSKGYTKSIDIWSVGCILAEMLSNRPIFPGKHYLDQLNHILGILGSPSQEDLNCIINLKARNYLLSLPHKNKVPWNRLFPNADSKALDLLDKMLTFNPHKRIEVEQALAHPYLEQYYDPSDEPIAEAPFKFDMELDDLPKEKLKELIFEETARFQPGYRS
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Serine/threonine kinase which acts as an essential component of the MAP kinase signal transduction pathway. MAPK1/ERK2 and MAPK3/ERK1 are the 2 MAPKs which play an important role in the MAPK/ERK cascade. They participate also in a signaling cascade initiated by activated KIT and KITLG/SCF. Depending on the cellular context, the MAPK/ERK cascade mediates diverse biological functions such as cell growth, adhesion, survival and differentiation through the regulation of transcription, translation, cytoskeletal rearrangements. The MAPK/ERK cascade plays also a role in initiation and regulation of meiosis, mitosis, and postmitotic functions in differentiated cells by phosphorylating a number of transcription factors. About 160 substrates have already been discovered for ERKs. Many of these substrates are localized in the nucleus, and seem to participate in the regulation of transcription upon stimulation. However, other substrates are found in the cytosol as well as in other cellular organelles, and those are responsible for processes such as translation, mitosis and apoptosis. Moreover, the MAPK/ERK cascade is also involved in the regulation of the endosomal dynamics, including lysosome processing and endosome cycling through the perinuclear recycling compartment (PNRC); as well as in the fragmentation of the Golgi apparatus during mitosis. The substrates include transcription factors (such as ATF2, BCL6, ELK1, ERF, FOS, HSF4 or SPZ1), cytoskeletal elements (such as CANX, CTTN, GJA1, MAP2, MAPT, PXN, SORBS3 or STMN1), regulators of apoptosis (such as BAD, BTG2, CASP9, DAPK1, IER3, MCL1 or PPARG), regulators of translation (such as EIF4EBP1) and a variety of other signaling-related molecules (like ARHGEF2, FRS2 or GRB10). Protein kinases (such as RAF1, RPS6KA1/RSK1, RPS6KA3/RSK2, RPS6KA2/RSK3, RPS6KA6/RSK4, SYK, MKNK1/MNK1, MKNK2/MNK2, RPS6KA5/MSK1, RPS6KA4/MSK2, MAPKAPK3 or MAPKAPK5) and phosphatases (such as DUSP1, DUSP4, DUSP6 or DUSP16) are other substrates which enable the propagation the MAPK/ERK signal to additional cytosolic and nuclear targets, thereby extending the specificity of the cascade.
Phosphorylated upon KIT and FLT3 signaling (By similarity). Dually phosphorylated on Thr-202 and Tyr-204, which activates the enzyme. Ligand-activated ALK induces tyrosine phosphorylation. Dephosphorylated by PTPRJ at Tyr-204.
Cytoplasm. Nucleus. Membrane>Caveola.
Note: Autophosphorylation at Thr-207 promotes nuclear localization.
The TXY motif contains the threonine and tyrosine residues whose phosphorylation activates the MAP kinases.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. MAP kinase subfamily.
Serine/threonine kinase which acts as an essential component of the MAP kinase signal transduction pathway. MAPK1/ERK2 and MAPK3/ERK1 are the 2 MAPKs which play an important role in the MAPK/ERK cascade. They participate also in a signaling cascade initiated by activated KIT and KITLG/SCF. Depending on the cellular context, the MAPK/ERK cascade mediates diverse biological functions such as cell growth, adhesion, survival and differentiation through the regulation of transcription, translation, cytoskeletal rearrangements. The MAPK/ERK cascade plays also a role in initiation and regulation of meiosis, mitosis, and postmitotic functions in differentiated cells by phosphorylating a number of transcription factors. About 160 substrates have already been discovered for ERKs. Many of these substrates are localized in the nucleus, and seem to participate in the regulation of transcription upon stimulation. However, other substrates are found in the cytosol as well as in other cellular organelles, and those are responsible for processes such as translation, mitosis and apoptosis. Moreover, the MAPK/ERK cascade is also involved in the regulation of the endosomal dynamics, including lysosome processing and endosome cycling through the perinuclear recycling compartment (PNRC); as well as in the fragmentation of the Golgi apparatus during mitosis. The substrates include transcription factors (such as ATF2, BCL6, ELK1, ERF, FOS, HSF4 or SPZ1), cytoskeletal elements (such as CANX, CTTN, GJA1, MAP2, MAPT, PXN, SORBS3 or STMN1), regulators of apoptosis (such as BAD, BTG2, CASP9, DAPK1, IER3, MCL1 or PPARG), regulators of translation (such as EIF4EBP1) and a variety of other signaling-related molecules (like ARHGEF2, DCC, FRS2 or GRB10). Protein kinases (such as RAF1, RPS6KA1/RSK1, RPS6KA3/RSK2, RPS6KA2/RSK3, RPS6KA6/RSK4, SYK, MKNK1/MNK1, MKNK2/MNK2, RPS6KA5/MSK1, RPS6KA4/MSK2, MAPKAPK3 or MAPKAPK5) and phosphatases (such as DUSP1, DUSP4, DUSP6 or DUSP16) are other substrates which enable the propagation the MAPK/ERK signal to additional cytosolic and nuclear targets, thereby extending the specificity of the cascade. Mediates phosphorylation of TPR in respons to EGF stimulation. May play a role in the spindle assembly checkpoint. Phosphorylates PML and promotes its interaction with PIN1, leading to PML degradation. Phosphorylates CDK2AP2 (By similarity).
Acts as a transcriptional repressor. Binds to a [GC]AAA[GC] consensus sequence. Repress the expression of interferon gamma-induced genes. Seems to bind to the promoter of CCL5, DMP1, IFIH1, IFITM1, IRF7, IRF9, LAMP3, OAS1, OAS2, OAS3 and STAT1. Transcriptional activity is independent of kinase activity.
Phosphorylated upon KIT and FLT3 signaling (By similarity). Dually phosphorylated on Thr-185 and Tyr-187, which activates the enzyme. Undergoes regulatory phosphorylation on additional residues such as Ser-246 and Ser-248 in the kinase insert domain (KID) These phosphorylations, which are probably mediated by more than one kinase, are important for binding of MAPK1/ERK2 to importin-7 (IPO7) and its nuclear translocation. In addition, autophosphorylation of Thr-190 was shown to affect the subcellular localization of MAPK1/ERK2 as well. Ligand-activated ALK induces tyrosine phosphorylation. Dephosphorylated by PTPRJ at Tyr-187. Phosphorylation on Ser-29 by SGK1 results in its activation by enhancing its interaction with MAP2K1/MEK1 and MAP2K2/MEK2. DUSP3 and DUSP6 dephosphorylate specifically MAPK1/ERK2 and MAPK3/ERK1 whereas DUSP9 dephosphorylates a broader range of MAPKs. Dephosphorylated by DUSP1 at Thr-185 and Tyr-187.
ISGylated.
Cytoplasm>Cytoskeleton>Spindle. Nucleus. Cytoplasm>Cytoskeleton>Microtubule organizing center>Centrosome. Cytoplasm. Membrane>Caveola.
Note: Associated with the spindle during prometaphase and metaphase (By similarity). PEA15-binding and phosphorylated DAPK1 promote its cytoplasmic retention. Phosphorylation at Ser- 246 and Ser-248 as well as autophosphorylation at Thr-190 promote nuclear localization.
The TXY motif contains the threonine and tyrosine residues whose phosphorylation activates the MAP kinases.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. MAP kinase subfamily.
Research Fields
· Cellular Processes > Cell growth and death > Oocyte meiosis. (View pathway)
· Cellular Processes > Transport and catabolism > Autophagy - animal. (View pathway)
· Cellular Processes > Cell growth and death > Apoptosis. (View pathway)
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Focal adhesion. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Adherens junction. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Gap junction. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Signaling pathways regulating pluripotency of stem cells. (View pathway)
· Cellular Processes > Cell motility > Regulation of actin cytoskeleton. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > ErbB signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Ras signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Rap1 signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cGMP-PKG signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cAMP signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > HIF-1 signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Sphingolipid signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Phospholipase D signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > mTOR signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > TGF-beta signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Apelin signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > TNF signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > EGFR tyrosine kinase inhibitor resistance.
· Human Diseases > Drug resistance: Antineoplastic > Endocrine resistance.
· Human Diseases > Drug resistance: Antineoplastic > Platinum drug resistance.
· Human Diseases > Endocrine and metabolic diseases > Type II diabetes mellitus.
· Human Diseases > Neurodegenerative diseases > Alzheimer's disease.
· Human Diseases > Neurodegenerative diseases > Prion diseases.
· Human Diseases > Substance dependence > Alcoholism.
· Human Diseases > Infectious diseases: Bacterial > Shigellosis.
· Human Diseases > Infectious diseases: Bacterial > Salmonella infection.
· Human Diseases > Infectious diseases: Bacterial > Pertussis.
· Human Diseases > Infectious diseases: Parasitic > Leishmaniasis.
· Human Diseases > Infectious diseases: Parasitic > Chagas disease (American trypanosomiasis).
· Human Diseases > Infectious diseases: Parasitic > Toxoplasmosis.
· Human Diseases > Infectious diseases: Bacterial > Tuberculosis.
· Human Diseases > Infectious diseases: Viral > Hepatitis C.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Influenza A.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Colorectal cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Renal cell carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Pancreatic cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Endometrial cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Glioma. (View pathway)
· Human Diseases > Cancers: Specific types > Prostate cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Thyroid cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Melanoma. (View pathway)
· Human Diseases > Cancers: Specific types > Bladder cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Chronic myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Acute myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Non-small cell lung cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Breast cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Hepatocellular carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Gastric cancer. (View pathway)
· Human Diseases > Cancers: Overview > Central carbon metabolism in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Choline metabolism in cancer. (View pathway)
· Organismal Systems > Immune system > Chemokine signaling pathway. (View pathway)
· Organismal Systems > Circulatory system > Adrenergic signaling in cardiomyocytes. (View pathway)
· Organismal Systems > Circulatory system > Vascular smooth muscle contraction. (View pathway)
· Organismal Systems > Development > Axon guidance. (View pathway)
· Organismal Systems > Development > Osteoclast differentiation. (View pathway)
· Organismal Systems > Immune system > Platelet activation. (View pathway)
· Organismal Systems > Immune system > Toll-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Natural killer cell mediated cytotoxicity. (View pathway)
· Organismal Systems > Immune system > IL-17 signaling pathway. (View pathway)
· Organismal Systems > Immune system > Th1 and Th2 cell differentiation. (View pathway)
· Organismal Systems > Immune system > Th17 cell differentiation. (View pathway)
· Organismal Systems > Immune system > T cell receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > B cell receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Fc epsilon RI signaling pathway. (View pathway)
· Organismal Systems > Immune system > Fc gamma R-mediated phagocytosis. (View pathway)
· Organismal Systems > Environmental adaptation > Circadian entrainment.
· Organismal Systems > Nervous system > Long-term potentiation.
· Organismal Systems > Nervous system > Neurotrophin signaling pathway. (View pathway)
· Organismal Systems > Nervous system > Retrograde endocannabinoid signaling. (View pathway)
· Organismal Systems > Nervous system > Glutamatergic synapse.
· Organismal Systems > Nervous system > Cholinergic synapse.
· Organismal Systems > Nervous system > Serotonergic synapse.
· Organismal Systems > Nervous system > Long-term depression.
· Organismal Systems > Endocrine system > Insulin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Progesterone-mediated oocyte maturation.
· Organismal Systems > Endocrine system > Estrogen signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Melanogenesis.
· Organismal Systems > Endocrine system > Prolactin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Thyroid hormone signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Oxytocin signaling pathway.
· Organismal Systems > Endocrine system > Relaxin signaling pathway.
· Organismal Systems > Excretory system > Aldosterone-regulated sodium reabsorption.
References
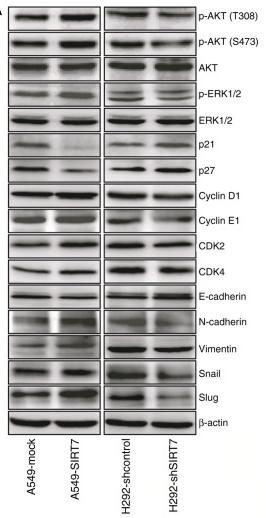
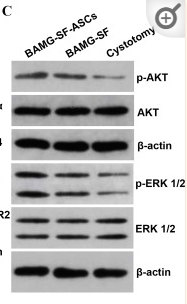
Application: WB Species: human Sample: myeloma cell
Application: WB Species: Rat Sample: rat aortic endothelial cells (RAECs)
Application: WB Species: Human Sample: NSCLC cells
Application: IHC Species: Human Sample: NSCLC cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.