Bcl-2 Antibody - #AF0769
| Product: | Bcl-2 Antibody |
| Catalog: | AF0769 |
| Description: | Rabbit polyclonal antibody to Bcl-2 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human |
| Mol.Wt.: | 28kDa; 26kD(Calculated). |
| Uniprot: | P10415 |
| RRID: | AB_2834113 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0769, RRID:AB_2834113.
Fold/Unfold
Apoptosis regulator Bcl 2; Apoptosis regulator Bcl-2; Apoptosis regulator Bcl2; AW986256; B cell CLL/lymphoma 2; B cell leukemia/lymphoma 2; Bcl-2; Bcl2; BCL2_HUMAN; C430015F12Rik; D630044D05Rik; D830018M01Rik; Leukemia/lymphoma, B-cell, 2; Oncogene B-cell leukemia 2; PPP1R50; Protein phosphatase 1, regulatory subunit 50;
Immunogens
A synthesized peptide derived from human Bcl-2, corresponding to a region within the internal amino acids.
- P10415 BCL2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAHAGRTGYDNREIVMKYIHYKLSQRGYEWDAGDVGAAPPGAAPAPGIFSSQPGHTPHPAASRDPVARTSPLQTPAAPGAAAGPALSPVPPVVHLTLRQAGDDFSRRYRRDFAEMSSQLHLTPFTARGRFATVVEELFRDGVNWGRIVAFFEFGGVMCVESVNREMSPLVDNIALWMTEYLNRHLHTWIQDNGGWDAFVELYGPSMRPLFDFSWLSLKTLLSLALVGACITLGAYLGHK
Research Backgrounds
Suppresses apoptosis in a variety of cell systems including factor-dependent lymphohematopoietic and neural cells. Regulates cell death by controlling the mitochondrial membrane permeability. Appears to function in a feedback loop system with caspases. Inhibits caspase activity either by preventing the release of cytochrome c from the mitochondria and/or by binding to the apoptosis-activating factor (APAF-1). May attenuate inflammation by impairing NLRP1-inflammasome activation, hence CASP1 activation and IL1B release.
Phosphorylation/dephosphorylation on Ser-70 regulates anti-apoptotic activity. Growth factor-stimulated phosphorylation on Ser-70 by PKC is required for the anti-apoptosis activity and occurs during the G2/M phase of the cell cycle. In the absence of growth factors, BCL2 appears to be phosphorylated by other protein kinases such as ERKs and stress-activated kinases. Phosphorylated by MAPK8/JNK1 at Thr-69, Ser-70 and Ser-87, wich stimulates starvation-induced autophagy. Dephosphorylated by protein phosphatase 2A (PP2A) (By similarity).
Proteolytically cleaved by caspases during apoptosis. The cleaved protein, lacking the BH4 motif, has pro-apoptotic activity, causes the release of cytochrome c into the cytosol promoting further caspase activity.
Monoubiquitinated by PRKN, leading to increase its stability. Ubiquitinated by SCF(FBXO10), leading to its degradation by the proteasome.
Mitochondrion outer membrane>Single-pass membrane protein. Nucleus membrane>Single-pass membrane protein. Endoplasmic reticulum membrane>Single-pass membrane protein.
Expressed in a variety of tissues.
BH1 and BH2 domains are required for the interaction with BAX and for anti-apoptotic activity.
The BH4 motif is required for anti-apoptotic activity and for interaction with RAF1 and EGLN3.
The loop between motifs BH4 and BH3 is required for the interaction with NLRP1.
Belongs to the Bcl-2 family.
Research Fields
· Cellular Processes > Transport and catabolism > Autophagy - animal. (View pathway)
· Cellular Processes > Cell growth and death > Apoptosis. (View pathway)
· Cellular Processes > Cell growth and death > Apoptosis - multiple species. (View pathway)
· Cellular Processes > Cell growth and death > Necroptosis. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Focal adhesion. (View pathway)
· Environmental Information Processing > Signal transduction > NF-kappa B signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > HIF-1 signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Sphingolipid signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hedgehog signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Jak-STAT signaling pathway. (View pathway)
· Genetic Information Processing > Folding, sorting and degradation > Protein processing in endoplasmic reticulum. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > EGFR tyrosine kinase inhibitor resistance.
· Human Diseases > Drug resistance: Antineoplastic > Endocrine resistance.
· Human Diseases > Drug resistance: Antineoplastic > Platinum drug resistance.
· Human Diseases > Neurodegenerative diseases > Amyotrophic lateral sclerosis (ALS).
· Human Diseases > Infectious diseases: Parasitic > Toxoplasmosis.
· Human Diseases > Infectious diseases: Bacterial > Tuberculosis.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Colorectal cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Prostate cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Small cell lung cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Gastric cancer. (View pathway)
· Organismal Systems > Circulatory system > Adrenergic signaling in cardiomyocytes. (View pathway)
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
· Organismal Systems > Nervous system > Neurotrophin signaling pathway. (View pathway)
· Organismal Systems > Nervous system > Cholinergic synapse.
· Organismal Systems > Endocrine system > Estrogen signaling pathway. (View pathway)
References
Application: WB Species: rat Sample: PC12 cells
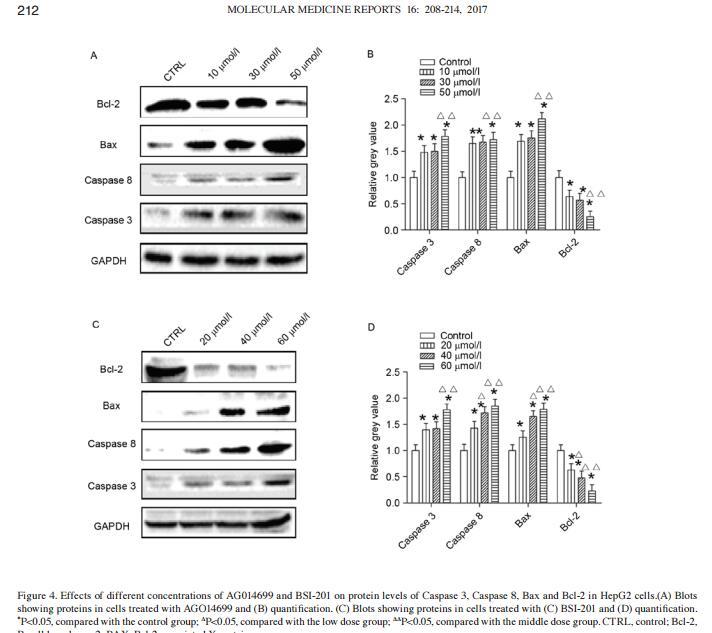
Application: WB Species: human Sample: HepG2
Application: WB Species: Human Sample: MG-63 cells
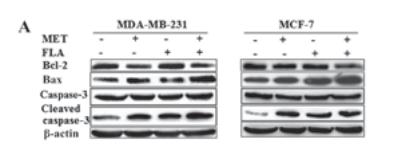
Application: WB Species: human Sample: MDA‑MB‑231 and MCF‑7 cells
Application: WB Species: Human Sample: AGS cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.