RBPJ Antibody - #DF7453
| Product: | RBPJ Antibody |
| Catalog: | DF7453 |
| Description: | Rabbit polyclonal antibody to RBPJ |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Sheep, Rabbit, Dog, Xenopus |
| Mol.Wt.: | 55kDa, 70 kDa; 56kD(Calculated). |
| Uniprot: | Q06330 |
| RRID: | AB_2839390 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF7453, RRID:AB_2839390.
Fold/Unfold
AI843960; AOS3; CBF 1; CBF-1; CBF1; csl; IGKJRB; IGKJRB1; J kappa recombination signal binding protein; J kappa-recombination signal-binding protein; KBF2; NY REN 30 antigen; RBP J; RBP J kappa; RBP JK; RBP-J; RBP-J kappa; RBP-JK; Rbpj; RBPJK; RBPSUH; recombination signal binding protein for immunoglobulin kappa J region; Recombining binding protein suppressor of hairless; Renal carcinoma antigen NY-REN-30; SUH; SUH_HUMAN;
Immunogens
A synthesized peptide derived from human RBPJ, corresponding to a region within C-terminal amino acids.
- Q06330 SUH_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MDHTEGSPAEEPPAHAPSPGKFGERPPPKRLTREAMRNYLKERGDQTVLILHAKVAQKSYGNEKRFFCPPPCVYLMGSGWKKKKEQMERDGCSEQESQPCAFIGIGNSDQEMQQLNLEGKNYCTAKTLYISDSDKRKHFMLSVKMFYGNSDDIGVFLSKRIKVISKPSKKKQSLKNADLCIASGTKVALFNRLRSQTVSTRYLHVEGGNFHASSQQWGAFFIHLLDDDESEGEEFTVRDGYIHYGQTVKLVCSVTGMALPRLIIRKVDKQTALLDADDPVSQLHKCAFYLKDTERMYLCLSQERIIQFQATPCPKEPNKEMINDGASWTIISTDKAEYTFYEGMGPVLAPVTPVPVVESLQLNGGGDVAMLELTGQNFTPNLRVWFGDVEAETMYRCGESMLCVVPDISAFREGWRWVRQPVQVPVTLVRNDGIIYSTSLTFTYTPEPGPRPHCSAAGAILRANSSQVPPNESNTNSEGSYTNASTNSTSVTSSTATVVS
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Transcriptional regulator that plays a central role in Notch signaling, a signaling pathway involved in cell-cell communication that regulates a broad spectrum of cell-fate determinations. Acts as a transcriptional repressor when it is not associated with Notch proteins. When associated with some NICD product of Notch proteins (Notch intracellular domain), it acts as a transcriptional activator that activates transcription of Notch target genes. Probably represses or activates transcription via the recruitment of chromatin remodeling complexes containing histone deacetylase or histone acetylase proteins, respectively. Specifically binds to the immunoglobulin kappa-type J segment recombination signal sequence. Binds specifically to methylated DNA. Binds to the oxygen responsive element of COX4I2 and activates its transcription under hypoxia conditions (4% oxygen). Negatively regulates the phagocyte oxidative burst in response to bacterial infection by repressing transcription of NADPH oxidase subunits (By similarity).
Nucleus. Cytoplasm.
Note: Mainly nuclear, upon interaction with RITA/C12orf52, translocates to the cytoplasm, down-regulating the Notch signaling pathway.
Belongs to the Su(H) family.
Research Fields
· Environmental Information Processing > Signal transduction > Notch signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Organismal Systems > Immune system > Th1 and Th2 cell differentiation. (View pathway)
References
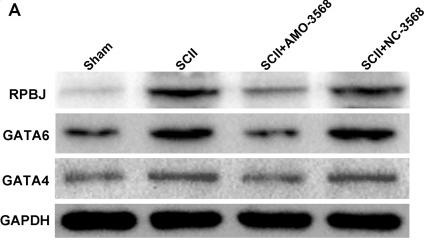
Application: WB Species: Rat Sample: spinal cord tissues
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.