NFAT2 Antibody - #DF6446
| Product: | NFAT2 Antibody |
| Catalog: | DF6446 |
| Description: | Rabbit polyclonal antibody to NFAT2 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Rabbit |
| Mol.Wt.: | 78,101kDa; 101kD(Calculated). |
| Uniprot: | O95644 |
| RRID: | AB_2838409 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF6446, RRID:AB_2838409.
Fold/Unfold
cytoplasmic 1; MGC138448; NF ATc; NF ATc1; NF-ATc; NF-ATc1; NF-ATc1.2; NFAC1_HUMAN; NFAT 2; NFAT transcription complex cytosolic component; NFATC 1; NFATc; NFATc1; Nuclear factor of activated T cells cytoplasmic 1; Nuclear factor of activated T cells cytoplasmic calcineurin dependent 1; Nuclear factor of activated T cells cytosolic component 1; nuclear factor of activated T-cells 'c'; Nuclear factor of activated T-cells;
Immunogens
A synthesized peptide derived from human NFAT2, corresponding to a region within the internal amino acids.
Expressed in thymus, peripheral leukocytes as T-cells and spleen. Isoforms A are preferentially expressed in effector T-cells (thymus and peripheral leukocytes) whereas isoforms B and isoforms C are preferentially expressed in naive T-cells (spleen). Isoforms B are expressed in naive T-cells after first antigen exposure and isoforms A are expressed in effector T-cells after second antigen exposure. Isoforms IA are widely expressed but not detected in liver nor pancreas, neural expression is strongest in corpus callosum. Isoforms IB are expressed mostly in muscle, cerebellum, placenta and thymus, neural expression in fetal and adult brain, strongest in corpus callosum.
- O95644 NFAC1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MPSTSFPVPSKFPLGPAAAVFGRGETLGPAPRAGGTMKSAEEEHYGYASSNVSPALPLPTAHSTLPAPCHNLQTSTPGIIPPADHPSGYGAALDGGPAGYFLSSGHTRPDGAPALESPRIEITSCLGLYHNNNQFFHDVEVEDVLPSSKRSPSTATLSLPSLEAYRDPSCLSPASSLSSRSCNSEASSYESNYSYPYASPQTSPWQSPCVSPKTTDPEEGFPRGLGACTLLGSPRHSPSTSPRASVTEESWLGARSSRPASPCNKRKYSLNGRQPPYSPHHSPTPSPHGSPRVSVTDDSWLGNTTQYTSSAIVAAINALTTDSSLDLGDGVPVKSRKTTLEQPPSVALKVEPVGEDLGSPPPPADFAPEDYSSFQHIRKGGFCDQYLAVPQHPYQWAKPKPLSPTSYMSPTLPALDWQLPSHSGPYELRIEVQPKSHHRAHYETEGSRGAVKASAGGHPIVQLHGYLENEPLMLQLFIGTADDRLLRPHAFYQVHRITGKTVSTTSHEAILSNTKVLEIPLLPENSMRAVIDCAGILKLRNSDIELRKGETDIGRKNTRVRLVFRVHVPQPSGRTLSLQVASNPIECSQRSAQELPLVEKQSTDSYPVVGGKKMVLSGHNFLQDSKVIFVEKAPDGHHVWEMEAKTDRDLCKPNSLVVEIPPFRNQRITSPVHVSFYVCNGKRKRSQYQRFTYLPANVPIIKTEPTDDYEPAPTCGPVSQGLSPLPRPYYSQQLAMPPDPSSCLVAGFPPCPQRSTLMPAAPGVSPKLHDLSPAAYTKGVASPGHCHLGLPQPAGEAPAVQDVPRPVATHPGSPGQPPPALLPQQVSAPPSSSCPPGLEHSLCPSSPSPPLPPATQEPTCLQPCSPACPPATGRPQHLPSTVRRDESPTAGPRLLPEVHEDGSPNLAPIPVTVKREPEELDQLYLDDVNEIIRNDLSSTSTHS
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Plays a role in the inducible expression of cytokine genes in T-cells, especially in the induction of the IL-2 or IL-4 gene transcription. Also controls gene expression in embryonic cardiac cells. Could regulate not only the activation and proliferation but also the differentiation and programmed death of T-lymphocytes as well as lymphoid and non-lymphoid cells. Required for osteoclastogenesis and regulates many genes important for osteoclast differentiation and function (By similarity).
Phosphorylated by NFATC-kinase and GSK3B; phosphorylation induces NFATC1 nuclear exit and dephosphorylation by calcineurin promotes nuclear import. Phosphorylation by PKA and DYRK2 negatively modulates nuclear accumulation, and promotes subsequent phosphorylation by GSK3B or casein kinase 1.
Cytoplasm. Nucleus.
Note: Cytoplasmic for the phosphorylated form and nuclear after activation that is controlled by calcineurin-mediated dephosphorylation. Rapid nuclear exit of NFATC is thought to be one mechanism by which cells distinguish between sustained and transient calcium signals. The subcellular localization of NFATC plays a key role in the regulation of gene transcription (PubMed:16511445). Nuclear translocation of NFATC1 is enhanced in the presence of TNFSF11. Nuclear translocation is decreased in the presence of FBN1 which can bind and sequester TNFSF11 (By similarity).
Expressed in thymus, peripheral leukocytes as T-cells and spleen. Isoforms A are preferentially expressed in effector T-cells (thymus and peripheral leukocytes) whereas isoforms B and isoforms C are preferentially expressed in naive T-cells (spleen). Isoforms B are expressed in naive T-cells after first antigen exposure and isoforms A are expressed in effector T-cells after second antigen exposure. Isoforms IA are widely expressed but not detected in liver nor pancreas, neural expression is strongest in corpus callosum. Isoforms IB are expressed mostly in muscle, cerebellum, placenta and thymus, neural expression in fetal and adult brain, strongest in corpus callosum.
Rel Similarity Domain (RSD) allows DNA-binding and cooperative interactions with AP1 factors.
The N-terminal transactivation domain (TAD-A) binds to and is activated by Cbp/p300. The dephosphorylated form contains two unmasked nuclear localization signals (NLS), which allow translocation of the protein to the nucleus.
Isoforms C have a C-terminal part with an additional trans-activation domain, TAD-B, which acts as a transcriptional activator. Isoforms B have a shorter C-terminal part without complete TAD-B which acts as a transcriptional repressor.
Research Fields
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cGMP-PKG signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cAMP signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Wnt signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Immune diseases > Inflammatory bowel disease (IBD).
· Organismal Systems > Development > Osteoclast differentiation. (View pathway)
· Organismal Systems > Immune system > Natural killer cell mediated cytotoxicity. (View pathway)
· Organismal Systems > Immune system > Th1 and Th2 cell differentiation. (View pathway)
· Organismal Systems > Immune system > Th17 cell differentiation. (View pathway)
· Organismal Systems > Immune system > T cell receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > B cell receptor signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Oxytocin signaling pathway.
References
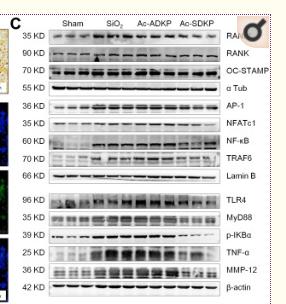
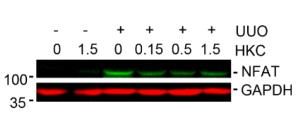
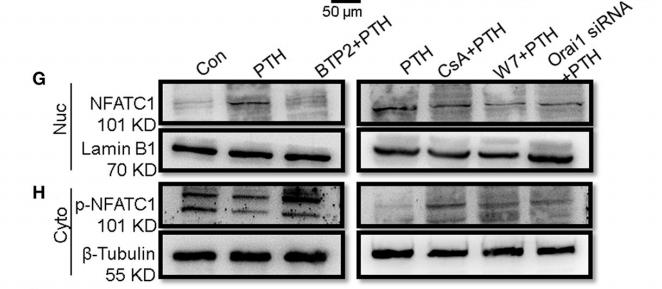
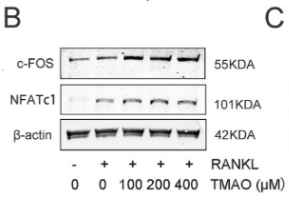
Application: IF/ICC Species: Mouse Sample: BMMs
Application: WB Species: Mouse Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.