Cleaved-Notch2 (Ala1734) Antibody - #AF5255
| Product: | Cleaved-Notch2 (Ala1734) Antibody |
| Catalog: | AF5255 |
| Description: | Rabbit polyclonal antibody to Cleaved-Notch2 (Ala1734) |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Dog, Chicken |
| Mol.Wt.: | 80kD(cleaved),300kD(FL); 265kD(Calculated). |
| Uniprot: | Q04721 |
| RRID: | AB_2837741 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF5255, RRID:AB_2837741.
Fold/Unfold
AGS2; hN2; Notch homolog 2; Notch2;
Immunogens
A synthesized peptide derived from human Notch2 (Cleaved-Ala1734).
Expressed in the brain, heart, kidney, lung, skeletal muscle and liver. Ubiquitously expressed in the embryo.
- Q04721 NOTC2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MPALRPALLWALLALWLCCAAPAHALQCRDGYEPCVNEGMCVTYHNGTGYCKCPEGFLGEYCQHRDPCEKNRCQNGGTCVAQAMLGKATCRCASGFTGEDCQYSTSHPCFVSRPCLNGGTCHMLSRDTYECTCQVGFTGKECQWTDACLSHPCANGSTCTTVANQFSCKCLTGFTGQKCETDVNECDIPGHCQHGGTCLNLPGSYQCQCPQGFTGQYCDSLYVPCAPSPCVNGGTCRQTGDFTFECNCLPGFEGSTCERNIDDCPNHRCQNGGVCVDGVNTYNCRCPPQWTGQFCTEDVDECLLQPNACQNGGTCANRNGGYGCVCVNGWSGDDCSENIDDCAFASCTPGSTCIDRVASFSCMCPEGKAGLLCHLDDACISNPCHKGALCDTNPLNGQYICTCPQGYKGADCTEDVDECAMANSNPCEHAGKCVNTDGAFHCECLKGYAGPRCEMDINECHSDPCQNDATCLDKIGGFTCLCMPGFKGVHCELEINECQSNPCVNNGQCVDKVNRFQCLCPPGFTGPVCQIDIDDCSSTPCLNGAKCIDHPNGYECQCATGFTGVLCEENIDNCDPDPCHHGQCQDGIDSYTCICNPGYMGAICSDQIDECYSSPCLNDGRCIDLVNGYQCNCQPGTSGVNCEINFDDCASNPCIHGICMDGINRYSCVCSPGFTGQRCNIDIDECASNPCRKGATCINGVNGFRCICPEGPHHPSCYSQVNECLSNPCIHGNCTGGLSGYKCLCDAGWVGINCEVDKNECLSNPCQNGGTCDNLVNGYRCTCKKGFKGYNCQVNIDECASNPCLNQGTCFDDISGYTCHCVLPYTGKNCQTVLAPCSPNPCENAAVCKESPNFESYTCLCAPGWQGQRCTIDIDECISKPCMNHGLCHNTQGSYMCECPPGFSGMDCEEDIDDCLANPCQNGGSCMDGVNTFSCLCLPGFTGDKCQTDMNECLSEPCKNGGTCSDYVNSYTCKCQAGFDGVHCENNINECTESSCFNGGTCVDGINSFSCLCPVGFTGSFCLHEINECSSHPCLNEGTCVDGLGTYRCSCPLGYTGKNCQTLVNLCSRSPCKNKGTCVQKKAESQCLCPSGWAGAYCDVPNVSCDIAASRRGVLVEHLCQHSGVCINAGNTHYCQCPLGYTGSYCEEQLDECASNPCQHGATCSDFIGGYRCECVPGYQGVNCEYEVDECQNQPCQNGGTCIDLVNHFKCSCPPGTRGLLCEENIDDCARGPHCLNGGQCMDRIGGYSCRCLPGFAGERCEGDINECLSNPCSSEGSLDCIQLTNDYLCVCRSAFTGRHCETFVDVCPQMPCLNGGTCAVASNMPDGFICRCPPGFSGARCQSSCGQVKCRKGEQCVHTASGPRCFCPSPRDCESGCASSPCQHGGSCHPQRQPPYYSCQCAPPFSGSRCELYTAPPSTPPATCLSQYCADKARDGVCDEACNSHACQWDGGDCSLTMENPWANCSSPLPCWDYINNQCDELCNTVECLFDNFECQGNSKTCKYDKYCADHFKDNHCDQGCNSEECGWDGLDCAADQPENLAEGTLVIVVLMPPEQLLQDARSFLRALGTLLHTNLRIKRDSQGELMVYPYYGEKSAAMKKQRMTRRSLPGEQEQEVAGSKVFLEIDNRQCVQDSDHCFKNTDAAAALLASHAIQGTLSYPLVSVVSESLTPERTQLLYLLAVAVVIILFIILLGVIMAKRKRKHGSLWLPEGFTLRRDASNHKRREPVGQDAVGLKNLSVQVSEANLIGTGTSEHWVDDEGPQPKKVKAEDEALLSEEDDPIDRRPWTQQHLEAADIRRTPSLALTPPQAEQEVDVLDVNVRGPDGCTPLMLASLRGGSSDLSDEDEDAEDSSANIITDLVYQGASLQAQTDRTGEMALHLAARYSRADAAKRLLDAGADANAQDNMGRCPLHAAVAADAQGVFQILIRNRVTDLDARMNDGTTPLILAARLAVEGMVAELINCQADVNAVDDHGKSALHWAAAVNNVEATLLLLKNGANRDMQDNKEETPLFLAAREGSYEAAKILLDHFANRDITDHMDRLPRDVARDRMHHDIVRLLDEYNVTPSPPGTVLTSALSPVICGPNRSFLSLKHTPMGKKSRRPSAKSTMPTSLPNLAKEAKDAKGSRRKKSLSEKVQLSESSVTLSPVDSLESPHTYVSDTTSSPMITSPGILQASPNPMLATAAPPAPVHAQHALSFSNLHEMQPLAHGASTVLPSVSQLLSHHHIVSPGSGSAGSLSRLHPVPVPADWMNRMEVNETQYNEMFGMVLAPAEGTHPGIAPQSRPPEGKHITTPREPLPPIVTFQLIPKGSIAQPAGAPQPQSTCPPAVAGPLPTMYQIPEMARLPSVAFPTAMMPQQDGQVAQTILPAYHPFPASVGKYPTPPSQHSYASSNAAERTPSHSGHLQGEHPYLTPSPESPDQWSSSSPHSASDWSDVTTSPTPGGAGGGQRGPGTHMSEPPHNNMQVYA
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity). Involved in bone remodeling and homeostasis. In collaboration with RELA/p65 enhances NFATc1 promoter activity and positively regulates RANKL-induced osteoclast differentiation. Positively regulates self-renewal of liver cancer cells.
Synthesized in the endoplasmic reticulum as an inactive form which is proteolytically cleaved by a furin-like convertase in the trans-Golgi network before it reaches the plasma membrane to yield an active, ligand-accessible form (By similarity). Cleavage results in a C-terminal fragment N(TM) and a N-terminal fragment N(EC) (By similarity). Following ligand binding, it is cleaved by TNF-alpha converting enzyme (TACE) to yield a membrane-associated intermediate fragment called notch extracellular truncation (NEXT) (By similarity). This fragment is then cleaved by presenilin dependent gamma-secretase to release a notch-derived peptide containing the intracellular domain (NICD) from the membrane (By similarity).
Hydroxylated by HIF1AN.
Can be either O-glucosylated or O-xylosylated at Ser-613 by POGLUT1.
Phosphorylated by GSK3. GSK3-mediated phosphorylation is necessary for NOTCH2 recognition by FBXW7, ubiquitination and degradation via the ubiquitin proteasome pathway.
Cell membrane>Single-pass type I membrane protein.
Nucleus. Cytoplasm.
Note: Following proteolytical processing NICD is translocated to the nucleus. Retained at the cytoplasm by TCIM (PubMed:25985737).
Expressed in the brain, heart, kidney, lung, skeletal muscle and liver. Ubiquitously expressed in the embryo.
Belongs to the NOTCH family.
Research Fields
· Environmental Information Processing > Signal transduction > Notch signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > Endocrine resistance.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Breast cancer. (View pathway)
· Organismal Systems > Immune system > Th1 and Th2 cell differentiation. (View pathway)
· Organismal Systems > Endocrine system > Thyroid hormone signaling pathway. (View pathway)
References
Application: WB Species: Mice Sample: oocytes and granular cells
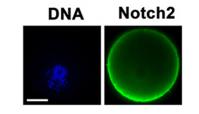
Application: IF/ICC Species: Mice Sample: oocytes and granular cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.