PI3K p85 alpha Antibody - #AF5121
| Product: | PI3K p85 alpha Antibody |
| Catalog: | AF5121 |
| Description: | Rabbit polyclonal antibody to PI3K p85 alpha |
| Application: | WB IHC |
| Cited expt.: | WB, IHC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 83 kDa; 84kD(Calculated). |
| Uniprot: | P27986 |
| RRID: | AB_2837607 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF5121, RRID:AB_2837607.
Fold/Unfold
GRB 1; GRB1; p85 alpha; p85; P85A_HUMAN; Phosphatidylinositol 3 kinase associated p 85 alpha; Phosphatidylinositol 3 kinase regulatory 1; Phosphatidylinositol 3 kinase regulatory subunit alpha; Phosphatidylinositol 3 kinase regulatory subunit polypeptide 1 (p85 alpha); Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha; Phosphatidylinositol 3-kinase regulatory subunit alpha; Phosphoinositide 3 kinase regulatory subunit 1 (alpha); Phosphoinositide 3 kinase regulatory subunit 1 (p85 alpha); Phosphoinositide 3 kinase regulatory subunit 1; Phosphoinositide 3 kinase regulatory subunit polypeptide 1 (p85 alpha); PI3 kinase p85 subunit alpha; PI3-kinase regulatory subunit alpha; PI3-kinase subunit p85-alpha; PI3K; PI3K regulatory subunit alpha; Pik3r1; PtdIns 3 kinase p85 alpha; PtdIns-3-kinase regulatory subunit alpha; PtdIns-3-kinase regulatory subunit p85-alpha;
Immunogens
A synthesized peptide derived from human PI3K p85 alpha, corresponding to a region within the internal amino acids.
Isoform 2 is expressed in skeletal muscle and brain, and at lower levels in kidney and cardiac muscle. Isoform 2 and isoform 4 are present in skeletal muscle (at protein level).
- P27986 P85A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSAEGYQYRALYDYKKEREEDIDLHLGDILTVNKGSLVALGFSDGQEARPEEIGWLNGYNETTGERGDFPGTYVEYIGRKKISPPTPKPRPPRPLPVAPGSSKTEADVEQQALTLPDLAEQFAPPDIAPPLLIKLVEAIEKKGLECSTLYRTQSSSNLAELRQLLDCDTPSVDLEMIDVHVLADAFKRYLLDLPNPVIPAAVYSEMISLAPEVQSSEEYIQLLKKLIRSPSIPHQYWLTLQYLLKHFFKLSQTSSKNLLNARVLSEIFSPMLFRFSAASSDNTENLIKVIEILISTEWNERQPAPALPPKPPKPTTVANNGMNNNMSLQDAEWYWGDISREEVNEKLRDTADGTFLVRDASTKMHGDYTLTLRKGGNNKLIKIFHRDGKYGFSDPLTFSSVVELINHYRNESLAQYNPKLDVKLLYPVSKYQQDQVVKEDNIEAVGKKLHEYNTQFQEKSREYDRLYEEYTRTSQEIQMKRTAIEAFNETIKIFEEQCQTQERYSKEYIEKFKREGNEKEIQRIMHNYDKLKSRISEIIDSRRRLEEDLKKQAAEYREIDKRMNSIKPDLIQLRKTRDQYLMWLTQKGVRQKKLNEWLGNENTEDQYSLVEDDEDLPHHDEKTWNVGSSNRNKAENLLRGKRDGTFLVRESSKQGCYACSVVVDGEVKHCVINKTATGYGFAEPYNLYSSLKELVLHYQHTSLVQHNDSLNVTLAYPVYAQQRR
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Binds to activated (phosphorylated) protein-Tyr kinases, through its SH2 domain, and acts as an adapter, mediating the association of the p110 catalytic unit to the plasma membrane. Necessary for the insulin-stimulated increase in glucose uptake and glycogen synthesis in insulin-sensitive tissues. Plays an important role in signaling in response to FGFR1, FGFR2, FGFR3, FGFR4, KITLG/SCF, KIT, PDGFRA and PDGFRB. Likewise, plays a role in ITGB2 signaling. Modulates the cellular response to ER stress by promoting nuclear translocation of XBP1 isoform 2 in a ER stress- and/or insulin-dependent manner during metabolic overloading in the liver and hence plays a role in glucose tolerance improvement.
Polyubiquitinated in T-cells by CBLB; which does not promote proteasomal degradation but impairs association with CD28 and CD3Z upon T-cell activation.
Phosphorylated. Tyrosine phosphorylated in response to signaling by FGFR1, FGFR2, FGFR3 and FGFR4. Phosphorylated by CSF1R. Phosphorylated by ERBB4. Phosphorylated on tyrosine residues by TEK/TIE2. Dephosphorylated by PTPRJ. Phosphorylated by PIK3CA at Ser-608; phosphorylation is stimulated by insulin and PDGF. The relevance of phosphorylation by PIK3CA is however unclear (By similarity). Phosphorylated in response to KIT and KITLG/SCF. Phosphorylated by FGR.
Isoform 2 is expressed in skeletal muscle and brain, and at lower levels in kidney and cardiac muscle. Isoform 2 and isoform 4 are present in skeletal muscle (at protein level).
The SH3 domain mediates the binding to CBLB, and to HIV-1 Nef.
Belongs to the PI3K p85 subunit family.
Research Fields
· Cellular Processes > Transport and catabolism > Autophagy - animal. (View pathway)
· Cellular Processes > Cell growth and death > Apoptosis. (View pathway)
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Focal adhesion. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Signaling pathways regulating pluripotency of stem cells. (View pathway)
· Cellular Processes > Cell motility > Regulation of actin cytoskeleton. (View pathway)
· Environmental Information Processing > Signal transduction > ErbB signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Ras signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Rap1 signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cAMP signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > HIF-1 signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Phosphatidylinositol signaling system.
· Environmental Information Processing > Signal transduction > Sphingolipid signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Phospholipase D signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > mTOR signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > AMPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Jak-STAT signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > TNF signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > EGFR tyrosine kinase inhibitor resistance.
· Human Diseases > Drug resistance: Antineoplastic > Endocrine resistance.
· Human Diseases > Drug resistance: Antineoplastic > Platinum drug resistance.
· Human Diseases > Endocrine and metabolic diseases > Type II diabetes mellitus.
· Human Diseases > Endocrine and metabolic diseases > Insulin resistance.
· Human Diseases > Endocrine and metabolic diseases > Non-alcoholic fatty liver disease (NAFLD).
· Human Diseases > Infectious diseases: Bacterial > Bacterial invasion of epithelial cells.
· Human Diseases > Infectious diseases: Parasitic > Chagas disease (American trypanosomiasis).
· Human Diseases > Infectious diseases: Parasitic > Amoebiasis.
· Human Diseases > Infectious diseases: Viral > Hepatitis C.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Measles.
· Human Diseases > Infectious diseases: Viral > Influenza A.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cancers: Specific types > Colorectal cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Renal cell carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Pancreatic cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Endometrial cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Glioma. (View pathway)
· Human Diseases > Cancers: Specific types > Prostate cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Melanoma. (View pathway)
· Human Diseases > Cancers: Specific types > Chronic myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Acute myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Small cell lung cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Non-small cell lung cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Breast cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Hepatocellular carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Gastric cancer. (View pathway)
· Human Diseases > Cancers: Overview > Central carbon metabolism in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Choline metabolism in cancer. (View pathway)
· Organismal Systems > Immune system > Chemokine signaling pathway. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway - multiple species. (View pathway)
· Organismal Systems > Development > Axon guidance. (View pathway)
· Organismal Systems > Development > Osteoclast differentiation. (View pathway)
· Organismal Systems > Immune system > Platelet activation. (View pathway)
· Organismal Systems > Immune system > Toll-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Natural killer cell mediated cytotoxicity. (View pathway)
· Organismal Systems > Immune system > T cell receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > B cell receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Fc epsilon RI signaling pathway. (View pathway)
· Organismal Systems > Immune system > Fc gamma R-mediated phagocytosis. (View pathway)
· Organismal Systems > Immune system > Leukocyte transendothelial migration. (View pathway)
· Organismal Systems > Nervous system > Neurotrophin signaling pathway. (View pathway)
· Organismal Systems > Nervous system > Cholinergic synapse.
· Organismal Systems > Sensory system > Inflammatory mediator regulation of TRP channels. (View pathway)
· Organismal Systems > Endocrine system > Insulin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Progesterone-mediated oocyte maturation.
· Organismal Systems > Endocrine system > Estrogen signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Prolactin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Thyroid hormone signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Regulation of lipolysis in adipocytes.
· Organismal Systems > Endocrine system > Relaxin signaling pathway.
· Organismal Systems > Excretory system > Aldosterone-regulated sodium reabsorption.
· Organismal Systems > Digestive system > Carbohydrate digestion and absorption.
References
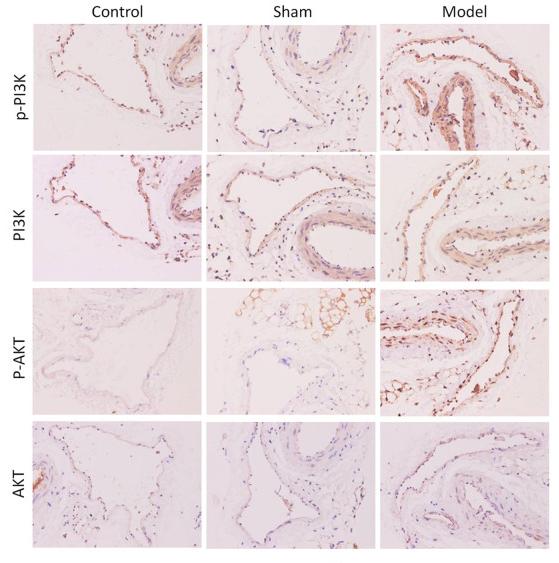
Application: IHC Species: Mice Sample: femoral vein tissue
Application: WB Species: Mice Sample: femoral vein tissue
Application: WB Species: Human Sample: breast carcinoma
Application: WB Species: Human Sample: AGS and MKN-45 cell
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.