IKK alpha/ beta Antibody - #AF6013
| Product: | IKK alpha/ beta Antibody |
| Catalog: | AF6013 |
| Description: | Rabbit polyclonal antibody to IKK alpha/ beta |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IHC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 85kDa; 85kD,87kD(Calculated). |
| Uniprot: | O15111 | O14920 |
| RRID: | AB_2834947 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF6013, RRID:AB_2834947.
Fold/Unfold
chuk; CHUK1; Conserved Helix Loop Helix Ubiquitous Kinase; Conserved helix loop ubiquitous kinase; Conserved helix-loop-helix ubiquitous kinase; I Kappa B Kinase 1; I Kappa B Kinase Alpha; I-kappa-B kinase 1; I-kappa-B kinase alpha; IkappaB kinase; IkB kinase alpha subunit; IkBKA; IKK 1; IKK A; IKK a kinase; IKK-A; IKK-alpha; IKK1; IKKA; IKKA_HUMAN; Inhibitor Of Kappa Light Polypeptide Gene Enhancer In B Cells; Inhibitor Of Nuclear Factor Kappa B Kinase Alpha Subunit; Inhibitor of nuclear factor kappa-B kinase subunit alpha; NFKBIKA; Nuclear Factor Kappa B Inhibitor Kinase Alpha; Nuclear factor NF kappa B inhibitor kinase alpha; Nuclear factor NF-kappa-B inhibitor kinase alpha; Nuclear factor NFkappaB inhibitor kinase alpha; Nuclear Factor Of Kappa Light Chain Gene Enhancer In B Cells Inhibitor; TCF-16; TCF16; Transcription factor 16; I kappa B kinase 2; I kappa B kinase beta; I-kappa-B kinase 2; I-kappa-B-kinase beta; IkBKB; IKK beta; IKK-B; IKK-beta; IKK2; IKKB; IKKB_HUMAN; IMD15; Inhibitor of kappa light polypeptide gene enhancer in B cells, kinase beta; Inhibitor of nuclear factor kappa-B kinase subunit beta; NFKBIKB; Nuclear factor NF-kappa-B inhibitor kinase beta;
Immunogens
A synthesized peptide derived from human IKK alpha/ beta, corresponding to a region within the internal amino acids.
Widely expressed.
O14920 IKKB_HUMAN:Highly expressed in heart, placenta, skeletal muscle, kidney, pancreas, spleen, thymus, prostate, testis and peripheral blood.
- O15111 IKKA_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MERPPGLRPGAGGPWEMRERLGTGGFGNVCLYQHRELDLKIAIKSCRLELSTKNRERWCHEIQIMKKLNHANVVKACDVPEELNILIHDVPLLAMEYCSGGDLRKLLNKPENCCGLKESQILSLLSDIGSGIRYLHENKIIHRDLKPENIVLQDVGGKIIHKIIDLGYAKDVDQGSLCTSFVGTLQYLAPELFENKPYTATVDYWSFGTMVFECIAGYRPFLHHLQPFTWHEKIKKKDPKCIFACEEMSGEVRFSSHLPQPNSLCSLVVEPMENWLQLMLNWDPQQRGGPVDLTLKQPRCFVLMDHILNLKIVHILNMTSAKIISFLLPPDESLHSLQSRIERETGINTGSQELLSETGISLDPRKPASQCVLDGVRGCDSYMVYLFDKSKTVYEGPFASRSLSDCVNYIVQDSKIQLPIIQLRKVWAEAVHYVSGLKEDYSRLFQGQRAAMLSLLRYNANLTKMKNTLISASQQLKAKLEFFHKSIQLDLERYSEQMTYGISSEKMLKAWKEMEEKAIHYAEVGVIGYLEDQIMSLHAEIMELQKSPYGRRQGDLMESLEQRAIDLYKQLKHRPSDHSYSDSTEMVKIIVHTVQSQDRVLKELFGHLSKLLGCKQKIIDLLPKVEVALSNIKEADNTVMFMQGKRQKEIWHLLKIACTQSSARSLVGSSLEGAVTPQTSAWLPPTSAEHDHSLSCVVTPQDGETSAQMIEENLNCLGHLSTIIHEANEEQGNSMMNLDWSWLTE
- O14920 IKKB_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSWSPSLTTQTCGAWEMKERLGTGGFGNVIRWHNQETGEQIAIKQCRQELSPRNRERWCLEIQIMRRLTHPNVVAARDVPEGMQNLAPNDLPLLAMEYCQGGDLRKYLNQFENCCGLREGAILTLLSDIASALRYLHENRIIHRDLKPENIVLQQGEQRLIHKIIDLGYAKELDQGSLCTSFVGTLQYLAPELLEQQKYTVTVDYWSFGTLAFECITGFRPFLPNWQPVQWHSKVRQKSEVDIVVSEDLNGTVKFSSSLPYPNNLNSVLAERLEKWLQLMLMWHPRQRGTDPTYGPNGCFKALDDILNLKLVHILNMVTGTIHTYPVTEDESLQSLKARIQQDTGIPEEDQELLQEAGLALIPDKPATQCISDGKLNEGHTLDMDLVFLFDNSKITYETQISPRPQPESVSCILQEPKRNLAFFQLRKVWGQVWHSIQTLKEDCNRLQQGQRAAMMNLLRNNSCLSKMKNSMASMSQQLKAKLDFFKTSIQIDLEKYSEQTEFGITSDKLLLAWREMEQAVELCGRENEVKLLVERMMALQTDIVDLQRSPMGRKQGGTLDDLEEQARELYRRLREKPRDQRTEGDSQEMVRLLLQAIQSFEKKVRVIYTQLSKTVVCKQKALELLPKVEEVVSLMNEDEKTVVRLQEKRQKELWNLLKIACSKVRGPVSGSPDSMNASRLSQPGQLMSQPSTASNSLPEPAKKSEELVAEAHNLCTLLENAIQDTVREQDQSFTALDWSWLQTEEEEHSCLEQAS
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Serine kinase that plays an essential role in the NF-kappa-B signaling pathway which is activated by multiple stimuli such as inflammatory cytokines, bacterial or viral products, DNA damages or other cellular stresses. Acts as part of the canonical IKK complex in the conventional pathway of NF-kappa-B activation and phosphorylates inhibitors of NF-kappa-B on serine residues. These modifications allow polyubiquitination of the inhibitors and subsequent degradation by the proteasome. In turn, free NF-kappa-B is translocated into the nucleus and activates the transcription of hundreds of genes involved in immune response, growth control, or protection against apoptosis. Negatively regulates the pathway by phosphorylating the scaffold protein TAXBP1 and thus promoting the assembly of the A20/TNFAIP3 ubiquitin-editing complex (composed of A20/TNFAIP3, TAX1BP1, and the E3 ligases ITCH and RNF11). Therefore, CHUK plays a key role in the negative feedback of NF-kappa-B canonical signaling to limit inflammatory gene activation. As part of the non-canonical pathway of NF-kappa-B activation, the MAP3K14-activated CHUK/IKKA homodimer phosphorylates NFKB2/p100 associated with RelB, inducing its proteolytic processing to NFKB2/p52 and the formation of NF-kappa-B RelB-p52 complexes. In turn, these complexes regulate genes encoding molecules involved in B-cell survival and lymphoid organogenesis. Participates also in the negative feedback of the non-canonical NF-kappa-B signaling pathway by phosphorylating and destabilizing MAP3K14/NIK. Within the nucleus, phosphorylates CREBBP and consequently increases both its transcriptional and histone acetyltransferase activities. Modulates chromatin accessibility at NF-kappa-B-responsive promoters by phosphorylating histones H3 at 'Ser-10' that are subsequently acetylated at 'Lys-14' by CREBBP. Additionally, phosphorylates the CREBBP-interacting protein NCOA3. Also phosphorylates FOXO3 and may regulate this pro-apoptotic transcription factor. Phosphorylates RIPK1 at 'Ser-25' which represses its kinase activity and consequently prevents TNF-mediated RIPK1-dependent cell death (By similarity).
Phosphorylated by MAP3K14/NIK, AKT and to a lesser extent by MEKK1, and dephosphorylated by PP2A. Autophosphorylated.
(Microbial infection) Acetylation of Thr-179 by Yersinia yopJ prevents phosphorylation and activation, thus blocking the I-kappa-B signaling pathway.
Cytoplasm. Nucleus.
Note: Shuttles between the cytoplasm and the nucleus.
Widely expressed.
The kinase domain is located in the N-terminal region. The leucine zipper is important to allow homo- and hetero-dimerization. At the C-terminal region is located the region responsible for the interaction with NEMO/IKBKG.
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family. I-kappa-B kinase subfamily.
Serine kinase that plays an essential role in the NF-kappa-B signaling pathway which is activated by multiple stimuli such as inflammatory cytokines, bacterial or viral products, DNA damages or other cellular stresses. Acts as part of the canonical IKK complex in the conventional pathway of NF-kappa-B activation. Phosphorylates inhibitors of NF-kappa-B on 2 critical serine residues. These modifications allow polyubiquitination of the inhibitors and subsequent degradation by the proteasome. In turn, free NF-kappa-B is translocated into the nucleus and activates the transcription of hundreds of genes involved in immune response, growth control, or protection against apoptosis. In addition to the NF-kappa-B inhibitors, phosphorylates several other components of the signaling pathway including NEMO/IKBKG, NF-kappa-B subunits RELA and NFKB1, as well as IKK-related kinases TBK1 and IKBKE. IKK-related kinase phosphorylations may prevent the overproduction of inflammatory mediators since they exert a negative regulation on canonical IKKs. Phosphorylates FOXO3, mediating the TNF-dependent inactivation of this pro-apoptotic transcription factor. Also phosphorylates other substrates including NCOA3, BCL10 and IRS1. Within the nucleus, acts as an adapter protein for NFKBIA degradation in UV-induced NF-kappa-B activation. Phosphorylates RIPK1 at 'Ser-25' which represses its kinase activity and consequently prevents TNF-mediated RIPK1-dependent cell death (By similarity).
Upon cytokine stimulation, phosphorylated on Ser-177 and Ser-181 by MEKK1 and/or MAP3K14/NIK as well as TBK1 and PRKCZ; which enhances activity. Once activated, autophosphorylates on the C-terminal serine cluster; which decreases activity and prevents prolonged activation of the inflammatory response. Phosphorylated by the IKK-related kinases TBK1 and IKBKE, which is associated with reduced CHUK/IKKA and IKBKB activity and NF-kappa-B-dependent gene transcription. Dephosphorylated at Ser-177 and Ser-181 by PPM1A and PPM1B.
(Microbial infection) Acetylation of Thr-180 by Yersinia yopJ prevents phosphorylation and activation, thus blocking the I-kappa-B pathway.
Ubiquitinated. Monoubiquitination involves TRIM21 that leads to inhibition of Tax-induced NF-kappa-B signaling. According to 'Ser-163' does not serve as a monoubiquitination site. According to ubiquitination on 'Ser-163' modulates phosphorylation on C-terminal serine residues.
(Microbial infection) Monoubiquitination by TRIM21 is disrupted by Yersinia yopJ.
Hydroxylated by PHD1/EGLN2, loss of hydroxylation under hypoxic conditions results in activation of NF-kappa-B.
Cytoplasm. Nucleus. Membrane raft.
Note: Colocalized with DPP4 in membrane rafts.
Highly expressed in heart, placenta, skeletal muscle, kidney, pancreas, spleen, thymus, prostate, testis and peripheral blood.
The kinase domain is located in the N-terminal region. The leucine zipper is important to allow homo- and hetero-dimerization. At the C-terminal region is located the region responsible for the interaction with NEMO/IKBKG.
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family. I-kappa-B kinase subfamily.
Research Fields
· Cellular Processes > Cell growth and death > Apoptosis. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Ras signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > NF-kappa B signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > mTOR signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > TNF signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > Antifolate resistance.
· Human Diseases > Endocrine and metabolic diseases > Type II diabetes mellitus.
· Human Diseases > Endocrine and metabolic diseases > Insulin resistance.
· Human Diseases > Endocrine and metabolic diseases > Non-alcoholic fatty liver disease (NAFLD).
· Human Diseases > Infectious diseases: Bacterial > Epithelial cell signaling in Helicobacter pylori infection.
· Human Diseases > Infectious diseases: Bacterial > Shigellosis.
· Human Diseases > Infectious diseases: Parasitic > Chagas disease (American trypanosomiasis).
· Human Diseases > Infectious diseases: Parasitic > Toxoplasmosis.
· Human Diseases > Infectious diseases: Viral > Hepatitis C.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Measles.
· Human Diseases > Infectious diseases: Viral > Influenza A.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Infectious diseases: Viral > Herpes simplex infection.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Pancreatic cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Prostate cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Chronic myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Acute myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Small cell lung cancer. (View pathway)
· Organismal Systems > Immune system > Chemokine signaling pathway. (View pathway)
· Organismal Systems > Development > Osteoclast differentiation. (View pathway)
· Organismal Systems > Immune system > Toll-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > RIG-I-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Cytosolic DNA-sensing pathway. (View pathway)
· Organismal Systems > Immune system > IL-17 signaling pathway. (View pathway)
· Organismal Systems > Immune system > Th1 and Th2 cell differentiation. (View pathway)
· Organismal Systems > Immune system > Th17 cell differentiation. (View pathway)
· Organismal Systems > Immune system > T cell receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > B cell receptor signaling pathway. (View pathway)
· Organismal Systems > Nervous system > Neurotrophin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Insulin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Adipocytokine signaling pathway.
References
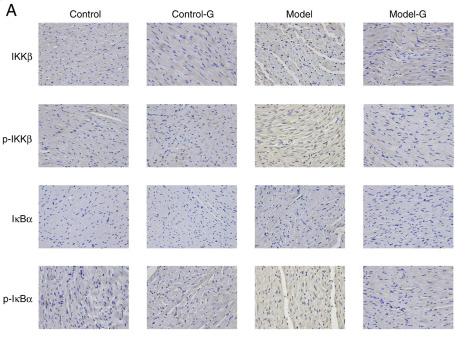
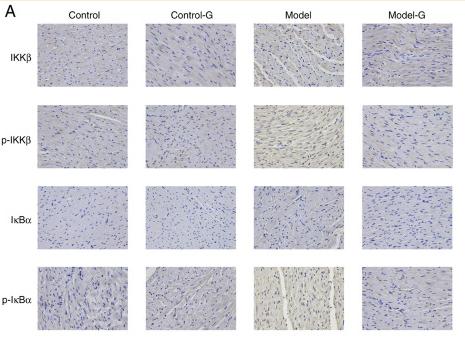
Application: IHC Species: Rat Sample: Heart tissues
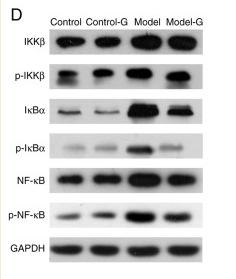
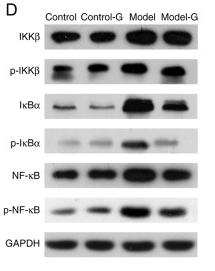
Application: WB Species: Rat Sample: Heart tissues
Application: IHC Species: Rat Sample: Heart tissues
Application: WB Species: Rat Sample: Heart tissues
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.