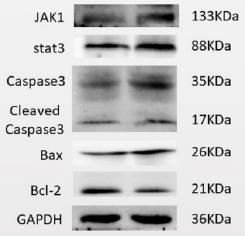
JAK1 Antibody - #AF5012
| Product: | JAK1 Antibody |
| Catalog: | AF5012 |
| Description: | Rabbit polyclonal antibody to JAK1 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat, Monkey |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 130kDa; 133kD(Calculated). |
| Uniprot: | P23458 |
| RRID: | AB_2834933 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF5012, RRID:AB_2834933.
Fold/Unfold
JAK 1; JAK 1A; JAK 1B; JAK-1; JAK1; JAK1_HUMAN; JAK1A; JAK1B; Janus kinase 1 (a protein tyrosine kinase); Janus kinase 1; JTK3; Tyrosine protein kinase JAK 1; Tyrosine protein kinase JAK1; Tyrosine-protein kinase JAK1;
Immunogens
A synthesized peptide derived from human JAK1, corresponding to a region within C-terminal amino acids.
Expressed at higher levels in primary colon tumors than in normal colon tissue. The expression level in metastatic colon tumors is comparable to the expression level in normal colon tissue.
- P23458 JAK1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MQYLNIKEDCNAMAFCAKMRSSKKTEVNLEAPEPGVEVIFYLSDREPLRLGSGEYTAEELCIRAAQACRISPLCHNLFALYDENTKLWYAPNRTITVDDKMSLRLHYRMRFYFTNWHGTNDNEQSVWRHSPKKQKNGYEKKKIPDATPLLDASSLEYLFAQGQYDLVKCLAPIRDPKTEQDGHDIENECLGMAVLAISHYAMMKKMQLPELPKDISYKRYIPETLNKSIRQRNLLTRMRINNVFKDFLKEFNNKTICDSSVSTHDLKVKYLATLETLTKHYGAEIFETSMLLISSENEMNWFHSNDGGNVLYYEVMVTGNLGIQWRHKPNVVSVEKEKNKLKRKKLENKHKKDEEKNKIREEWNNFSYFPEITHIVIKESVVSINKQDNKKMELKLSSHEEALSFVSLVDGYFRLTADAHHYLCTDVAPPLIVHNIQNGCHGPICTEYAINKLRQEGSEEGMYVLRWSCTDFDNILMTVTCFEKSEQVQGAQKQFKNFQIEVQKGRYSLHGSDRSFPSLGDLMSHLKKQILRTDNISFMLKRCCQPKPREISNLLVATKKAQEWQPVYPMSQLSFDRILKKDLVQGEHLGRGTRTHIYSGTLMDYKDDEGTSEEKKIKVILKVLDPSHRDISLAFFEAASMMRQVSHKHIVYLYGVCVRDVENIMVEEFVEGGPLDLFMHRKSDVLTTPWKFKVAKQLASALSYLEDKDLVHGNVCTKNLLLAREGIDSECGPFIKLSDPGIPITVLSRQECIERIPWIAPECVEDSKNLSVAADKWSFGTTLWEICYNGEIPLKDKTLIEKERFYESRCRPVTPSCKELADLMTRCMNYDPNQRPFFRAIMRDINKLEEQNPDIVSEKKPATEVDPTHFEKRFLKRIRDLGEGHFGKVELCRYDPEGDNTGEQVAVKSLKPESGGNHIADLKKEIEILRNLYHENIVKYKGICTEDGGNGIKLIMEFLPSGSLKEYLPKNKNKINLKQQLKYAVQICKGMDYLGSRQYVHRDLAARNVLVESEHQVKIGDFGLTKAIETDKEYYTVKDDRDSPVFWYAPECLMQSKFYIASDVWSFGVTLHELLTYCDSDSSPMALFLKMIGPTHGQMTVTRLVNTLKEGKRLPCPPNCPDEVYQLMRKCWEFQPSNRTSFQNLIEGFEALLK
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Tyrosine kinase of the non-receptor type, involved in the IFN-alpha/beta/gamma signal pathway. Kinase partner for the interleukin (IL)-2 receptor as well as interleukin (IL)-10 receptor.
Autophosphorylated. Phosphorylated on tyrosine residues in response to interferon gamma signaling. Dephosphorylation of Tyr-1034 and Tyr-1035 by PTPN2 negatively regulates cytokine-mediated signaling.
Ubiquitinated by RNF125; leading to its degradation by the proteasome.
Endomembrane system>Peripheral membrane protein.
Note: Wholly intracellular, possibly membrane associated.
Expressed at higher levels in primary colon tumors than in normal colon tissue. The expression level in metastatic colon tumors is comparable to the expression level in normal colon tissue.
Possesses two phosphotransferase domains. The second one probably contains the catalytic domain, while the presence of slight differences suggest a different role for domain 1.
The FERM domain mediates interaction with JAKMIP1.
Belongs to the protein kinase superfamily. Tyr protein kinase family. JAK subfamily.
Research Fields
· Cellular Processes > Cell growth and death > Necroptosis. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Signaling pathways regulating pluripotency of stem cells. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Jak-STAT signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > EGFR tyrosine kinase inhibitor resistance.
· Human Diseases > Infectious diseases: Parasitic > Leishmaniasis.
· Human Diseases > Infectious diseases: Parasitic > Toxoplasmosis.
· Human Diseases > Infectious diseases: Bacterial > Tuberculosis.
· Human Diseases > Infectious diseases: Viral > Hepatitis C.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Measles.
· Human Diseases > Infectious diseases: Viral > Influenza A.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Infectious diseases: Viral > Herpes simplex infection.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Human Diseases > Cancers: Specific types > Pancreatic cancer. (View pathway)
· Organismal Systems > Development > Osteoclast differentiation. (View pathway)
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Th1 and Th2 cell differentiation. (View pathway)
· Organismal Systems > Immune system > Th17 cell differentiation. (View pathway)
References
Application: WB Species: Mice Sample:
Application: WB Species: Fish Sample: zebrafish larvae
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.





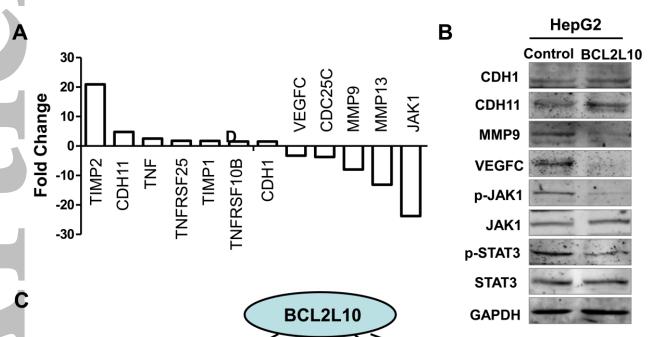
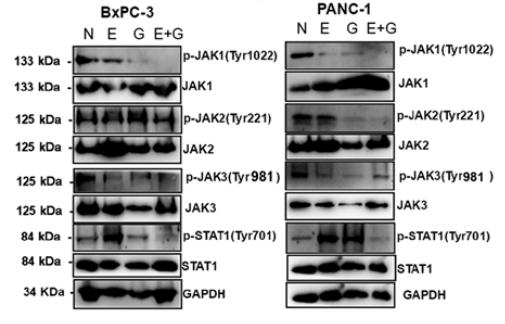
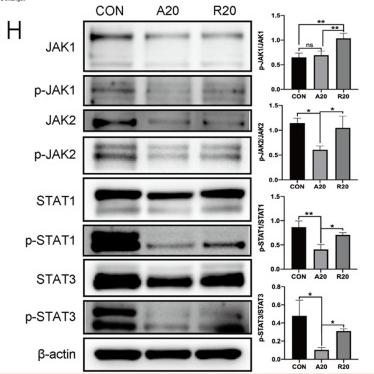
![Figure 5
A‐F, The expression of JAK1, p‐JAK1, JAK2, p‐JAK2, STAT1, and p‐STAT1 in macrophages within each group as determined by western blot analysis. G‐I, Quantitative analysis of the bands measured by gray value. The experiment was carried out a minimum of three times, and values are expressed as the mean ± SD. *P < .05 and **P < .01 compared with MI. JAK, Janus kinase; STAT, signal transducer and activator of transcription [Color figure can be viewed at wileyonlinelibrary.com] JAK1 Antibody - Figure 5
A‐F, The expression of JAK1, p‐JAK1, JAK2, p‐JAK2, STAT1, and p‐STAT1 in macrophages within each group as determined by western blot analysis.](http://img.affbiotech.cn/images/cited_image/202207/cite-wx-66-1658977738.jpg)