Phospho-STAT3 (Ser727) Antibody - #AF3294
| Product: | Phospho-STAT3 (Ser727) Antibody |
| Catalog: | AF3294 |
| Description: | Rabbit polyclonal antibody to Phospho-STAT3 (Ser727) |
| Application: | WB IHC IF/ICC IP |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Chicken |
| Mol.Wt.: | 86kDa; 88kD(Calculated). |
| Uniprot: | P40763 |
| RRID: | AB_2834713 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF3294, RRID:AB_2834713.
Fold/Unfold
1110034C02Rik; Acute Phase Response Factor; Acute-phase response factor; ADMIO; APRF; AW109958; DNA binding protein APRF; FLJ20882; HIES; MGC16063; Signal transducer and activator of transcription 3 (acute phase response factor); Signal transducer and activator of transcription 3; STAT 3; Stat3; STAT3_HUMAN;
Immunogens
A synthesized peptide derived from human STAT3 around the phosphorylation site of Ser727.
- P40763 STAT3_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAQWNQLQQLDTRYLEQLHQLYSDSFPMELRQFLAPWIESQDWAYAASKESHATLVFHNLLGEIDQQYSRFLQESNVLYQHNLRRIKQFLQSRYLEKPMEIARIVARCLWEESRLLQTAATAAQQGGQANHPTAAVVTEKQQMLEQHLQDVRKRVQDLEQKMKVVENLQDDFDFNYKTLKSQGDMQDLNGNNQSVTRQKMQQLEQMLTALDQMRRSIVSELAGLLSAMEYVQKTLTDEELADWKRRQQIACIGGPPNICLDRLENWITSLAESQLQTRQQIKKLEELQQKVSYKGDPIVQHRPMLEERIVELFRNLMKSAFVVERQPCMPMHPDRPLVIKTGVQFTTKVRLLVKFPELNYQLKIKVCIDKDSGDVAALRGSRKFNILGTNTKVMNMEESNNGSLSAEFKHLTLREQRCGNGGRANCDASLIVTEELHLITFETEVYHQGLKIDLETHSLPVVVISNICQMPNAWASILWYNMLTNNPKNVNFFTKPPIGTWDQVAEVLSWQFSSTTKRGLSIEQLTTLAEKLLGPGVNYSGCQITWAKFCKENMAGKGFSFWVWLDNIIDLVKKYILALWNEGYIMGFISKERERAILSTKPPGTFLLRFSESSKEGGVTFTWVEKDISGKTQIQSVEPYTKQQLNNMSFAEIIMGYKIMDATNILVSPLVYLYPDIPKEEAFGKYCRPESQEHPEADPGSAAPYLKTKFICVTPTTCSNTIDLPMSPRTLDSLMQFGNNGEGAEPSAGGQFESLTFDMELTSECATSPM
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Signal transducer and transcription activator that mediates cellular responses to interleukins, KITLG/SCF, LEP and other growth factors. Once activated, recruits coactivators, such as NCOA1 or MED1, to the promoter region of the target gene. May mediate cellular responses to activated FGFR1, FGFR2, FGFR3 and FGFR4. Binds to the interleukin-6 (IL-6)-responsive elements identified in the promoters of various acute-phase protein genes. Activated by IL31 through IL31RA. Acts as a regulator of inflammatory response by regulating differentiation of naive CD4(+) T-cells into T-helper Th17 or regulatory T-cells (Treg): deacetylation and oxidation of lysine residues by LOXL3, leads to disrupt STAT3 dimerization and inhibit its transcription activity. Involved in cell cycle regulation by inducing the expression of key genes for the progression from G1 to S phase, such as CCND1. Mediates the effects of LEP on melanocortin production, body energy homeostasis and lactation (By similarity). May play an apoptotic role by transctivating BIRC5 expression under LEP activation. Cytoplasmic STAT3 represses macroautophagy by inhibiting EIF2AK2/PKR activity. Plays a crucial role in basal beta cell functions, such as regulation of insulin secretion (By similarity).
Tyrosine phosphorylated upon stimulation with EGF. Tyrosine phosphorylated in response to constitutively activated FGFR1, FGFR2, FGFR3 and FGFR4 (By similarity). Activated through tyrosine phosphorylation by BMX. Tyrosine phosphorylated in response to IL6, IL11, LIF, CNTF, KITLG/SCF, CSF1, EGF, PDGF, IFN-alpha, LEP and OSM. Activated KIT promotes phosphorylation on tyrosine residues and subsequent translocation to the nucleus. Phosphorylated on serine upon DNA damage, probably by ATM or ATR. Serine phosphorylation is important for the formation of stable DNA-binding STAT3 homodimers and maximal transcriptional activity. ARL2BP may participate in keeping the phosphorylated state of STAT3 within the nucleus. Upon LPS challenge, phosphorylated within the nucleus by IRAK1. Upon erythropoietin treatment, phosphorylated on Ser-727 by RPS6KA5. Phosphorylation at Tyr-705 by PTK6 or FER leads to an increase of its transcriptional activity. Dephosphorylation on tyrosine residues by PTPN2 negatively regulates IL6/interleukin-6 signaling.
Acetylated on lysine residues by CREBBP. Deacetylation by LOXL3 leads to disrupt STAT3 dimerization and inhibit STAT3 transcription activity. Oxidation of lysine residues to allysine on STAT3 preferentially takes place on lysine residues that are acetylated.
Some lysine residues are oxidized to allysine by LOXL3, leading to disrupt STAT3 dimerization and inhibit STAT3 transcription activity. Oxidation of lysine residues to allysine on STAT3 preferentially takes place on lysine residues that are acetylated.
(Microbial infection) Phosphorylated on Tyr-705 in the presence of S.typhimurium SarA.
S-palmitoylated by ZDHHC19 in SH2 putative lipid-binding pockets, leading to homodimerization. Nuclear STAT3 is highly palmitoylated (about 75%) compared with cytoplasmic STAT3 (about 20%).
S-stearoylated, probably by ZDHHC19.
Cytoplasm. Nucleus.
Note: Shuttles between the nucleus and the cytoplasm. Translocated into the nucleus upon tyrosine phosphorylation and dimerization, in response to signaling by activated FGFR1, FGFR2, FGFR3 or FGFR4. Constitutive nuclear presence is independent of tyrosine phosphorylation. Predominantly present in the cytoplasm without stimuli. Upon leukemia inhibitory factor (LIF) stimulation, accumulates in the nucleus. The complex composed of BART and ARL2 plays an important role in the nuclear translocation and retention of STAT3. Identified in a complex with LYN and PAG1.
Heart, brain, placenta, lung, liver, skeletal muscle, kidney and pancreas.
Belongs to the transcription factor STAT family.
Research Fields
· Cellular Processes > Cell growth and death > Necroptosis. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Signaling pathways regulating pluripotency of stem cells. (View pathway)
· Environmental Information Processing > Signal transduction > HIF-1 signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Jak-STAT signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > EGFR tyrosine kinase inhibitor resistance.
· Human Diseases > Endocrine and metabolic diseases > Insulin resistance.
· Human Diseases > Infectious diseases: Parasitic > Toxoplasmosis.
· Human Diseases > Infectious diseases: Viral > Hepatitis C.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Measles.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Pancreatic cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Acute myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Non-small cell lung cancer. (View pathway)
· Human Diseases > Immune diseases > Inflammatory bowel disease (IBD).
· Organismal Systems > Immune system > Chemokine signaling pathway. (View pathway)
· Organismal Systems > Immune system > Th17 cell differentiation. (View pathway)
· Organismal Systems > Endocrine system > Prolactin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Adipocytokine signaling pathway.
References
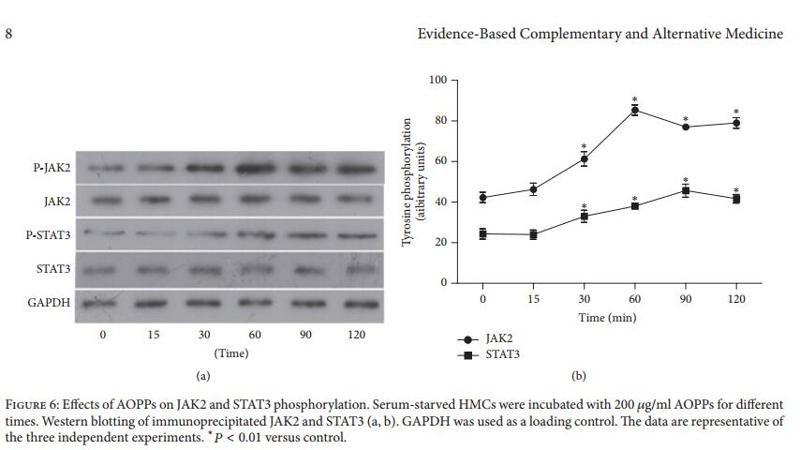
Application: WB Species: Mice Sample: EpH4-Ev cells
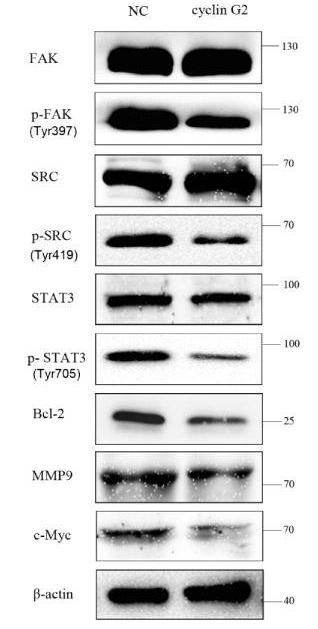
Application: WB Species: Rat Sample:
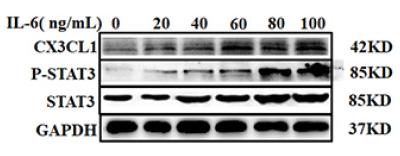
Application: WB Species: Human Sample: SCC-9 cells
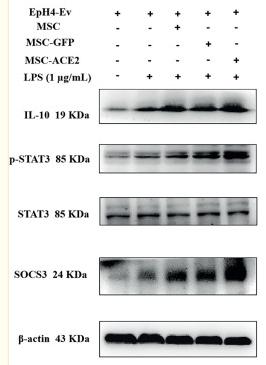
Application: WB Species: Human Sample: HUVECs
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.