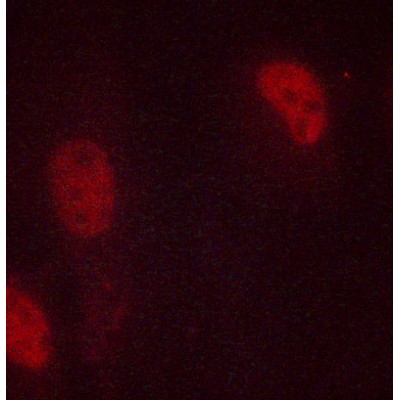
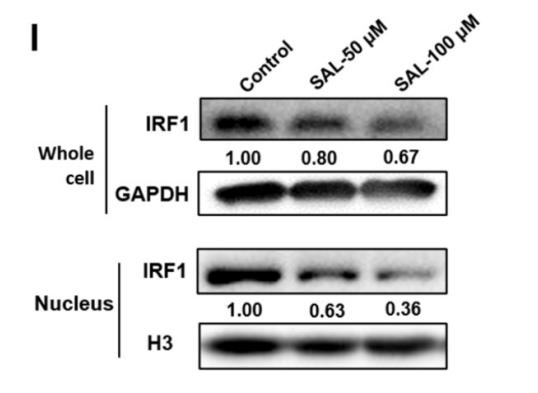
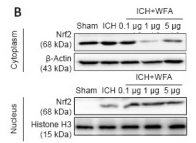
Histone H3 Antibody - #AF0863
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0863, RRID:AB_2810277.
Fold/Unfold
H3 histone family, member A; H3/A; H31_HUMAN; H3FA; Hist1h3a; HIST1H3B; HIST1H3C; HIST1H3D; HIST1H3E; HIST1H3F; HIST1H3G; HIST1H3H; HIST1H3I; HIST1H3J; histone 1, H3a; Histone cluster 1, H3a; Histone H3.1; Histone H3/a; Histone H3/b; Histone H3/c; Histone H3/d; Histone H3/f; Histone H3/h; Histone H3/i; Histone H3/j; Histone H3/k; Histone H3/l; ;H3.3A; HIST1 cluster, H3E; H3 histone family, member A; H3.1; H3/l; H3F3; H3FF; H3FJ; H3FL; Histone gene cluster 1, H3 histone family, member E; histone H3.1t; Histone H3/o; FLJ92264; H 3; H3; H3 histone family, member B; H3 histone family, member C; H3 histone family, member D; H3 histone family, member F; H3 histone family, member H; H3 histone family, member I; H3 histone family, member J; H3 histone family, member K; H3 histone family, member L; H3 histone family, member T; H3 histone, family 3A; H3/A; H3/b; H3/c; H3/d; h3/f; H3/h; H3/i; H3/j; H3/k; H3/t; H31_HUMAN; H3F1K; H3F3A; H3FA; H3FB; H3FC; H3FD; H3FH; H3FI; H3FK; HIST1 cluster, H3A; HIST1 cluster, H3B; HIST1 cluster, H3C; HIST1 cluster, H3D; HIST1 cluster, H3F; HIST1 cluster, H3G; HIST1 cluster, H3H; HIST1 cluster, H3I; HIST1 cluster, H3J; HIST1H3A; HIST1H3B; HIST1H3C; HIST1H3D; HIST1H3E; HIST1H3F; HIST1H3G; HIST1H3H; HIST1H3I; HIST1H3J; HIST3H3; Histone 1, H3a; Histone 1, H3b; Histone 1, H3c; Histone 1, H3d; Histone 1, H3e; Histone 1, H3f; Histone 1, H3g; Histone 1, H3h; Histone 1, H3i; Histone 3, H3; histone cluster 1 H3 family member a; histone cluster 1 H3 family member b; histone cluster 1 H3 family member c; histone cluster 1 H3 family member d; histone cluster 1 H3 family member e; histone cluster 1 H3 family member f; histone cluster 1 H3 family member g; histone cluster 1 H3 family member h; histone cluster 1 H3 family member i; histone cluster 1 H3 family member j; Histone cluster 1, H3a; Histone cluster 1, H3b; Histone cluster 1, H3c; Histone cluster 1, H3d; Histone cluster 1, H3e; Histone cluster 1, H3f; Histone cluster 1, H3g; Histone cluster 1, H3i; Histone cluster 1, H3j; Histone gene cluster 1, H3 histone family, member A; Histone gene cluster 1, H3 histone family, member B; Histone gene cluster 1, H3 histone family, member C; Histone gene cluster 1, H3 histone family, member D; Histone gene cluster 1, H3 histone family, member F; Histone gene cluster 1, H3 histone family, member G; Histone gene cluster 1, H3 histone family, member H; Histone gene cluster 1, H3 histone family, member I; Histone gene cluster 1, H3 histone family, member J; Histone gene cluster 1, H3A; Histone gene cluster 1, H3B; Histone gene cluster 1, H3C; Histone gene cluster 1, H3D; Histone gene cluster 1, H3E; Histone gene cluster 1, H3F; Histone gene cluster 1, H3G; Histone gene cluster 1, H3H; Histone gene cluster 1, H3I; Histone gene cluster 1, H3J; Histone H 3; Histone H3.1; Histone H3.2; Histone H3.3; Histone H3/a; Histone H3/b; Histone H3/c; Histone H3/d; Histone H3/f; Histone H3/h; Histone H3/i; Histone H3/j; Histone H3/k; Histone H3/l; Histone H3/m; H3 histone family 3A; H3 histone family 3B; H3 histone, family 3B (H3.3B); H3.3; H3.3A; H3.3B; H33_HUMAN; H3F3; H3F3A; H3f3b; Histone H3.3; Histone H3.3Q; Histone H3.A; Histone H3.B; MGC87782; MGC87783;
Immunogens
A synthesized peptide derived from human Histone H3.
- P68431 H31_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPHRYRPGTVALREIRRYQKSTELLIRKLPFQRLVREIAQDFKTDLRFQSSAVMALQEACEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA
- Q71DI3 H32_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPHRYRPGTVALREIRRYQKSTELLIRKLPFQRLVREIAQDFKTDLRFQSSAVMALQEASEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA
- P84243 H33_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MARTKQTARKSTGGKAPRKQLATKAARKSAPSTGGVKKPHRYRPGTVALREIRRYQKSTELLIRKLPFQRLVREIAQDFKTDLRFQSAAIGALQEASEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Core component of nucleosome. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling.
Acetylation is generally linked to gene activation. Acetylation on Lys-10 (H3K9ac) impairs methylation at Arg-9 (H3R8me2s). Acetylation on Lys-19 (H3K18ac) and Lys-24 (H3K24ac) favors methylation at Arg-18 (H3R17me). Acetylation at Lys-123 (H3K122ac) by EP300/p300 plays a central role in chromatin structure: localizes at the surface of the histone octamer and stimulates transcription, possibly by promoting nucleosome instability.
Citrullination at Arg-9 (H3R8ci) and/or Arg-18 (H3R17ci) by PADI4 impairs methylation and represses transcription.
Asymmetric dimethylation at Arg-18 (H3R17me2a) by CARM1 is linked to gene activation. Symmetric dimethylation at Arg-9 (H3R8me2s) by PRMT5 is linked to gene repression. Asymmetric dimethylation at Arg-3 (H3R2me2a) by PRMT6 is linked to gene repression and is mutually exclusive with H3 Lys-5 methylation (H3K4me2 and H3K4me3). H3R2me2a is present at the 3' of genes regardless of their transcription state and is enriched on inactive promoters, while it is absent on active promoters.
Methylation at Lys-5 (H3K4me), Lys-37 (H3K36me) and Lys-80 (H3K79me) are linked to gene activation. Methylation at Lys-5 (H3K4me) facilitates subsequent acetylation of H3 and H4. Methylation at Lys-80 (H3K79me) is associated with DNA double-strand break (DSB) responses and is a specific target for TP53BP1. Methylation at Lys-10 (H3K9me) and Lys-28 (H3K27me) are linked to gene repression. Methylation at Lys-10 (H3K9me) is a specific target for HP1 proteins (CBX1, CBX3 and CBX5) and prevents subsequent phosphorylation at Ser-11 (H3S10ph) and acetylation of H3 and H4. Methylation at Lys-5 (H3K4me) and Lys-80 (H3K79me) require preliminary monoubiquitination of H2B at 'Lys-120'. Methylation at Lys-10 (H3K9me) and Lys-28 (H3K27me) are enriched in inactive X chromosome chromatin. Monomethylation at Lys-57 (H3K56me1) by EHMT2/G9A in G1 phase promotes interaction with PCNA and is required for DNA replication.
Phosphorylated at Thr-4 (H3T3ph) by HASPIN during prophase and dephosphorylated during anaphase. Phosphorylation at Ser-11 (H3S10ph) by AURKB is crucial for chromosome condensation and cell-cycle progression during mitosis and meiosis. In addition phosphorylation at Ser-11 (H3S10ph) by RPS6KA4 and RPS6KA5 is important during interphase because it enables the transcription of genes following external stimulation, like mitogens, stress, growth factors or UV irradiation and result in the activation of genes, such as c-fos and c-jun. Phosphorylation at Ser-11 (H3S10ph), which is linked to gene activation, prevents methylation at Lys-10 (H3K9me) but facilitates acetylation of H3 and H4. Phosphorylation at Ser-11 (H3S10ph) by AURKB mediates the dissociation of HP1 proteins (CBX1, CBX3 and CBX5) from heterochromatin. Phosphorylation at Ser-11 (H3S10ph) is also an essential regulatory mechanism for neoplastic cell transformation. Phosphorylated at Ser-29 (H3S28ph) by MAP3K20 isoform 1, RPS6KA5 or AURKB during mitosis or upon ultraviolet B irradiation. Phosphorylation at Thr-7 (H3T6ph) by PRKCB is a specific tag for epigenetic transcriptional activation that prevents demethylation of Lys-5 (H3K4me) by LSD1/KDM1A. At centromeres, specifically phosphorylated at Thr-12 (H3T11ph) from prophase to early anaphase, by DAPK3 and PKN1. Phosphorylation at Thr-12 (H3T11ph) by PKN1 is a specific tag for epigenetic transcriptional activation that promotes demethylation of Lys-10 (H3K9me) by KDM4C/JMJD2C. Phosphorylation at Thr-12 (H3T11ph) by chromatin-associated CHEK1 regulates the transcription of cell cycle regulatory genes by modulating acetylation of Lys-10 (H3K9ac). Phosphorylation at Tyr-42 (H3Y41ph) by JAK2 promotes exclusion of CBX5 (HP1 alpha) from chromatin.
Monoubiquitinated by RAG1 in lymphoid cells, monoubiquitination is required for V(D)J recombination (By similarity). Ubiquitinated by the CUL4-DDB-RBX1 complex in response to ultraviolet irradiation. This may weaken the interaction between histones and DNA and facilitate DNA accessibility to repair proteins.
Lysine deamination at Lys-5 (H3K4all) to form allysine is mediated by LOXL2. Allysine formation by LOXL2 only takes place on H3K4me3 and results in gene repression.
Crotonylation (Kcr) is specifically present in male germ cells and marks testis-specific genes in post-meiotic cells, including X-linked genes that escape sex chromosome inactivation in haploid cells. Crotonylation marks active promoters and enhancers and confers resistance to transcriptional repressors. It is also associated with post-meiotically activated genes on autosomes.
Butyrylation of histones marks active promoters and competes with histone acetylation. It is present during late spermatogenesis.
Succinylation at Lys-80 (H3K79succ) by KAT2A takes place with a maximum frequency around the transcription start sites of genes. It gives a specific tag for epigenetic transcription activation. Desuccinylation at Lys-123 (H3K122succ) by SIRT7 in response to DNA damage promotes chromatin condensation and double-strand breaks (DSBs) repair.
Serine ADP-ribosylation constitutes the primary form of ADP-ribosylation of proteins in response to DNA damage. Serine ADP-ribosylation at Ser-11 (H3S10ADPr) is mutually exclusive with phosphorylation at Ser-11 (H3S10ph) and impairs acetylation at Lys-10 (H3K9ac).
Nucleus. Chromosome.
Belongs to the histone H3 family.
Core component of nucleosome. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling.
Acetylation is generally linked to gene activation. Acetylation on Lys-10 (H3K9ac) impairs methylation at Arg-9 (H3R8me2s). Acetylation on Lys-19 (H3K18ac) and Lys-24 (H3K24ac) favors methylation at Arg-18 (H3R17me). Acetylation at Lys-123 (H3K122ac) by EP300/p300 plays a central role in chromatin structure: localizes at the surface of the histone octamer and stimulates transcription, possibly by promoting nucleosome instability.
Citrullination at Arg-9 (H3R8ci) and/or Arg-18 (H3R17ci) by PADI4 impairs methylation and represses transcription.
Asymmetric dimethylation at Arg-18 (H3R17me2a) by CARM1 is linked to gene activation. Symmetric dimethylation at Arg-9 (H3R8me2s) by PRMT5 is linked to gene repression. Asymmetric dimethylation at Arg-3 (H3R2me2a) by PRMT6 is linked to gene repression and is mutually exclusive with H3 Lys-5 methylation (H3K4me2 and H3K4me3). H3R2me2a is present at the 3' of genes regardless of their transcription state and is enriched on inactive promoters, while it is absent on active promoters.
Methylation at Lys-5 (H3K4me), Lys-37 (H3K36me) and Lys-80 (H3K79me) are linked to gene activation. Methylation at Lys-5 (H3K4me) facilitates subsequent acetylation of H3 and H4. Methylation at Lys-80 (H3K79me) is associated with DNA double-strand break (DSB) responses and is a specific target for TP53BP1. Methylation at Lys-10 (H3K9me) and Lys-28 (H3K27me) are linked to gene repression. Methylation at Lys-10 (H3K9me) is a specific target for HP1 proteins (CBX1, CBX3 and CBX5) and prevents subsequent phosphorylation at Ser-11 (H3S10ph) and acetylation of H3 and H4. Methylation at Lys-5 (H3K4me) and Lys-80 (H3K79me) require preliminary monoubiquitination of H2B at 'Lys-120'. Methylation at Lys-10 (H3K9me) and Lys-28 (H3K27me) are enriched in inactive X chromosome chromatin. Monomethylation at Lys-57 (H3K56me1) by EHMT2/G9A in G1 phase promotes interaction with PCNA and is required for DNA replication.
Phosphorylated at Thr-4 (H3T3ph) by HASPIN during prophase and dephosphorylated during anaphase. Phosphorylation at Ser-11 (H3S10ph) by AURKB is crucial for chromosome condensation and cell-cycle progression during mitosis and meiosis. In addition phosphorylation at Ser-11 (H3S10ph) by RPS6KA4 and RPS6KA5 is important during interphase because it enables the transcription of genes following external stimulation, like mitogens, stress, growth factors or UV irradiation and result in the activation of genes, such as c-fos and c-jun. Phosphorylation at Ser-11 (H3S10ph), which is linked to gene activation, prevents methylation at Lys-10 (H3K9me) but facilitates acetylation of H3 and H4. Phosphorylation at Ser-11 (H3S10ph) by AURKB mediates the dissociation of HP1 proteins (CBX1, CBX3 and CBX5) from heterochromatin. Phosphorylation at Ser-11 (H3S10ph) is also an essential regulatory mechanism for neoplastic cell transformation. Phosphorylated at Ser-29 (H3S28ph) by MAP3K20 isoform 1, RPS6KA5 or AURKB during mitosis or upon ultraviolet B irradiation. Phosphorylation at Thr-7 (H3T6ph) by PRKCB is a specific tag for epigenetic transcriptional activation that prevents demethylation of Lys-5 (H3K4me) by LSD1/KDM1A. At centromeres, specifically phosphorylated at Thr-12 (H3T11ph) from prophase to early anaphase, by DAPK3 and PKN1. Phosphorylation at Thr-12 (H3T11ph) by PKN1 is a specific tag for epigenetic transcriptional activation that promotes demethylation of Lys-10 (H3K9me) by KDM4C/JMJD2C. Phosphorylation at Tyr-42 (H3Y41ph) by JAK2 promotes exclusion of CBX5 (HP1 alpha) from chromatin.
Monoubiquitinated by RAG1 in lymphoid cells, monoubiquitination is required for V(D)J recombination. Ubiquitinated by the CUL4-DDB-RBX1 complex in response to ultraviolet irradiation. This may weaken the interaction between histones and DNA and facilitate DNA accessibility to repair proteins.
Lysine deamination at Lys-5 (H3K4all) to form allysine is mediated by LOXL2. Allysine formation by LOXL2 only takes place on H3K4me3 and results in gene repression.
Crotonylation (Kcr) is specifically present in male germ cells and marks testis-specific genes in post-meiotic cells, including X-linked genes that escape sex chromosome inactivation in haploid cells. Crotonylation marks active promoters and enhancers and confers resistance to transcriptional repressors. It is also associated with post-meiotically activated genes on autosomes.
Butyrylation of histones marks active promoters and competes with histone acetylation. It is present during late spermatogenesis.
Succinylation at Lys-80 (H3K79succ) by KAT2A takes place with a maximum frequency around the transcription start sites of genes. It gives a specific tag for epigenetic transcription activation. Desuccinylation at Lys-123 (H3K122succ) by SIRT7 in response to DNA damage promotes chromatin condensation and double-strand breaks (DSBs) repair.
Serine ADP-ribosylation constitutes the primary form of ADP-ribosylation of proteins in response to DNA damage. Serine ADP-ribosylation at Ser-11 (H3S10ADPr) is mutually exclusive with phosphorylation at Ser-11 (H3S10ph) and impairs acetylation at Lys-10 (H3K9ac).
Nucleus. Chromosome.
Belongs to the histone H3 family.
Variant histone H3 which replaces conventional H3 in a wide range of nucleosomes in active genes. Constitutes the predominant form of histone H3 in non-dividing cells and is incorporated into chromatin independently of DNA synthesis. Deposited at sites of nucleosomal displacement throughout transcribed genes, suggesting that it represents an epigenetic imprint of transcriptionally active chromatin. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling.
Acetylation is generally linked to gene activation. Acetylation on Lys-10 (H3K9ac) impairs methylation at Arg-9 (H3R8me2s). Acetylation on Lys-19 (H3K18ac) and Lys-24 (H3K24ac) favors methylation at Arg-18 (H3R17me). Acetylation at Lys-123 (H3K122ac) by EP300/p300 plays a central role in chromatin structure: localizes at the surface of the histone octamer and stimulates transcription, possibly by promoting nucleosome instability.
Citrullination at Arg-9 (H3R8ci) and/or Arg-18 (H3R17ci) by PADI4 impairs methylation and represses transcription.
Asymmetric dimethylation at Arg-18 (H3R17me2a) by CARM1 is linked to gene activation. Symmetric dimethylation at Arg-9 (H3R8me2s) by PRMT5 is linked to gene repression. Asymmetric dimethylation at Arg-3 (H3R2me2a) by PRMT6 is linked to gene repression and is mutually exclusive with H3 Lys-5 methylation (H3K4me2 and H3K4me3). H3R2me2a is present at the 3' of genes regardless of their transcription state and is enriched on inactive promoters, while it is absent on active promoters.
Specifically enriched in modifications associated with active chromatin such as methylation at Lys-5 (H3K4me), Lys-37 and Lys-80. Methylation at Lys-5 (H3K4me) facilitates subsequent acetylation of H3 and H4. Methylation at Lys-80 (H3K79me) is associated with DNA double-strand break (DSB) responses and is a specific target for TP53BP1. Methylation at Lys-10 (H3K9me) and Lys-28 (H3K27me), which are linked to gene repression, are underrepresented. Methylation at Lys-10 (H3K9me) is a specific target for HP1 proteins (CBX1, CBX3 and CBX5) and prevents subsequent phosphorylation at Ser-11 (H3S10ph) and acetylation of H3 and H4. Methylation at Lys-5 (H3K4me) and Lys-80 (H3K79me) require preliminary monoubiquitination of H2B at 'Lys-120'. Methylation at Lys-10 (H3K9me) and Lys-28 (H3K27me) are enriched in inactive X chromosome chromatin. Monomethylation at Lys-57 (H3K56me1) by EHMT2/G9A in G1 phase promotes interaction with PCNA and is required for DNA replication.
Phosphorylated at Thr-4 (H3T3ph) by HASPIN during prophase and dephosphorylated during anaphase. Phosphorylation at Ser-11 (H3S10ph) by AURKB is crucial for chromosome condensation and cell-cycle progression during mitosis and meiosis. In addition phosphorylation at Ser-11 (H3S10ph) by RPS6KA4 and RPS6KA5 is important during interphase because it enables the transcription of genes following external stimulation, like mitogens, stress, growth factors or UV irradiation and result in the activation of genes, such as c-fos and c-jun. Phosphorylation at Ser-11 (H3S10ph), which is linked to gene activation, prevents methylation at Lys-10 (H3K9me) but facilitates acetylation of H3 and H4. Phosphorylation at Ser-11 (H3S10ph) by AURKB mediates the dissociation of HP1 proteins (CBX1, CBX3 and CBX5) from heterochromatin. Phosphorylation at Ser-11 (H3S10ph) is also an essential regulatory mechanism for neoplastic cell transformation. Phosphorylated at Ser-29 (H3S28ph) by MAP3K20 isoform 1, RPS6KA5 or AURKB during mitosis or upon ultraviolet B irradiation. Phosphorylation at Thr-7 (H3T6ph) by PRKCB is a specific tag for epigenetic transcriptional activation that prevents demethylation of Lys-5 (H3K4me) by LSD1/KDM1A. At centromeres, specifically phosphorylated at Thr-12 (H3T11ph) from prophase to early anaphase, by DAPK3 and PKN1. Phosphorylation at Thr-12 (H3T11ph) by PKN1 is a specific tag for epigenetic transcriptional activation that promotes demethylation of Lys-10 (H3K9me) by KDM4C/JMJD2C. Phosphorylation at Tyr-42 (H3Y41ph) by JAK2 promotes exclusion of CBX5 (HP1 alpha) from chromatin. Phosphorylation on Ser-32 (H3S31ph) is specific to regions bordering centromeres in metaphase chromosomes.
Ubiquitinated. Monoubiquitinated by RAG1 in lymphoid cells, monoubiquitination is required for V(D)J recombination (By similarity).
Lysine deamination at Lys-5 (H3K4all) to form allysine is mediated by LOXL2. Allysine formation by LOXL2 only takes place on H3K4me3 and results in gene repression.
Crotonylation (Kcr) is specifically present in male germ cells and marks testis-specific genes in post-meiotic cells, including X-linked genes that escape sex chromosome inactivation in haploid cells. Crotonylation marks active promoters and enhancers and confers resistance to transcriptional repressors. It is also associated with post-meiotically activated genes on autosomes.
Butyrylation of histones marks active promoters and competes with histone acetylation. It is present during late spermatogenesis.
Succinylation at Lys-80 (H3K79succ) by KAT2A takes place with a maximum frequency around the transcription start sites of genes. It gives a specific tag for epigenetic transcription activation. Desuccinylation at Lys-123 (H3K122succ) by SIRT7 in response to DNA damage promotes chromatin condensation and double-strand breaks (DSBs) repair.
Serine ADP-ribosylation constitutes the primary form of ADP-ribosylation of proteins in response to DNA damage. Serine ADP-ribosylation at Ser-11 (H3S10ADPr) is mutually exclusive with phosphorylation at Ser-11 (H3S10ph) and impairs acetylation at Lys-10 (H3K9ac).
Nucleus. Chromosome.
Specific interaction of trimethylated form at 'Lys-36' (H3.3K36me3) with ZMYND11 is mediated by the encapsulation of Ser-32 residue with a composite pocket formed by the tandem bromo-PWWP domains.
Belongs to the histone H3 family.
Research Fields
· Human Diseases > Substance dependence > Alcoholism.
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
· Human Diseases > Immune diseases > Systemic lupus erythematosus.
References
Application: WB Species: mouse Sample: B16F10 and 4T1 tumor
Application: WB Species: human Sample: A375 cells
Application: WB Species: Mouse Sample:
Application: WB Species: Mice Sample: colonic tissues
Application: WB Species: Mice Sample: kidney tissue
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.