TBK1 Antibody - #DF7026
| Product: | TBK1 Antibody |
| Catalog: | DF7026 |
| Description: | Rabbit polyclonal antibody to TBK1 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 84kDa; 84kD(Calculated). |
| Uniprot: | Q9UHD2 |
| RRID: | AB_2838982 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF7026, RRID:AB_2838982.
Fold/Unfold
EC 2.7.11.1; FLJ11330; FTDALS4; NAK; NF kappa B activating kinase; NF kB activating kinase; NF-kappa-B-activating kinase; Serine/threonine protein kinase TBK 1; Serine/threonine protein kinase TBK1; Serine/threonine-protein kinase TBK1; T2K; TANK binding kinase 1; TANK-binding kinase 1; TBK 1; Tbk1; TBK1_HUMAN;
Immunogens
A synthesized peptide derived from human TBK1, corresponding to a region within the internal amino acids.
Ubiquitous with higher expression in testis. Expressed in the ganglion cells, nerve fiber layer and microvasculature of the retina.
- Q9UHD2 TBK1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MQSTSNHLWLLSDILGQGATANVFRGRHKKTGDLFAIKVFNNISFLRPVDVQMREFEVLKKLNHKNIVKLFAIEEETTTRHKVLIMEFCPCGSLYTVLEEPSNAYGLPESEFLIVLRDVVGGMNHLRENGIVHRDIKPGNIMRVIGEDGQSVYKLTDFGAARELEDDEQFVSLYGTEEYLHPDMYERAVLRKDHQKKYGATVDLWSIGVTFYHAATGSLPFRPFEGPRRNKEVMYKIITGKPSGAISGVQKAENGPIDWSGDMPVSCSLSRGLQVLLTPVLANILEADQEKCWGFDQFFAETSDILHRMVIHVFSLQQMTAHKIYIHSYNTATIFHELVYKQTKIISSNQELIYEGRRLVLEPGRLAQHFPKTTEENPIFVVSREPLNTIGLIYEKISLPKVHPRYDLDGDASMAKAITGVVCYACRIASTLLLYQELMRKGIRWLIELIKDDYNETVHKKTEVVITLDFCIRNIEKTVKVYEKLMKINLEAAELGEISDIHTKLLRLSSSQGTIETSLQDIDSRLSPGGSLADAWAHQEGTHPKDRNVEKLQVLLNCMTEIYYQFKKDKAERRLAYNEEQIHKFDKQKLYYHATKAMTHFTDECVKKYEAFLNKSEEWIRKMLHLRKQLLSLTNQCFDIEEEVSKYQEYTNELQETLPQKMFTASSGIKHTMTPIYPSSNTLVEMTLGMKKLKEEMEGVVKELAENNHILERFGSLTMDGGLRNVDCL
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Serine/threonine kinase that plays an essential role in regulating inflammatory responses to foreign agents. Following activation of toll-like receptors by viral or bacterial components, associates with TRAF3 and TANK and phosphorylates interferon regulatory factors (IRFs) IRF3 and IRF7 as well as DDX3X. This activity allows subsequent homodimerization and nuclear translocation of the IRFs leading to transcriptional activation of pro-inflammatory and antiviral genes including IFNA and IFNB. In order to establish such an antiviral state, TBK1 form several different complexes whose composition depends on the type of cell and cellular stimuli. Plays a key role in IRF3 activation: acts by first phosphorylating innate adapter proteins MAVS, STING1 and TICAM1 on their pLxIS motif, leading to recruitment of IRF3, thereby licensing IRF3 for phosphorylation by TBK1. Phosphorylated IRF3 dissociates from the adapter proteins, dimerizes, and then enters the nucleus to induce expression of interferons. Thus, several scaffolding molecules including FADD, TRADD, MAVS, AZI2, TANK or TBKBP1/SINTBAD can be recruited to the TBK1-containing-complexes. Under particular conditions, functions as a NF-kappa-B effector by phosphorylating NF-kappa-B inhibitor alpha/NFKBIA, IKBKB or RELA to translocate NF-Kappa-B to the nucleus. Restricts bacterial proliferation by phosphorylating the autophagy receptor OPTN/Optineurin on 'Ser-177', thus enhancing LC3 binding affinity and antibacterial autophagy. Phosphorylates SMCR8 component of the C9orf72-SMCR8 complex, promoting autophagosome maturation. Phosphorylates and activates AKT1. Seems to play a role in energy balance regulation by sustaining a state of chronic, low-grade inflammation in obesity, wich leads to a negative impact on insulin sensitivity (By similarity). Attenuates retroviral budding by phosphorylating the endosomal sorting complex required for transport-I (ESCRT-I) subunit VPS37C. Phosphorylates Borna disease virus (BDV) P protein. Plays an essential role in the TLR3- and IFN-dependent control of herpes virus HSV-1 and HSV-2 infections in the central nervous system.
Autophosphorylation at Ser-172 activates the kinase, and is an essential step for virus-triggered signaling. Phosphorylated by IKBKB/IKKB at Ser-172. Phosphorylation requires homodimerization and ubiquitination at Lys-30 and Lys-401. Dephosphorylated at Ser-172 by PPM1B and this negatively regulates its role in mediating antiviral response.
'Lys-63'-linked polyubiquitination by MIB1 after RNA virus infection, or by NRDP1 after LPS stimulation at Lys-30 and Lys-401, participates in kinase activation. 'Lys-48'-linked polyubiquitination at Lys-670 by DTX4 leads to proteasomal degradation. 'Lys-48'-linked polyubiquitination by TRAIP also leads to proteasomal degradation. 'Lys-63'-linked polyubiquitination by RNF128 at Lys-30 and Lys-401 leads to the activation of antiviral responses.
Cytoplasm.
Note: Upon mitogen stimulation or triggering of the immune system, TBK1 is recruited to the exocyst by EXOC2.
Ubiquitous with higher expression in testis. Expressed in the ganglion cells, nerve fiber layer and microvasculature of the retina.
Comprises A N-terminal kinase domain, a ubiquitin-like domain and a C-terminal coiled-coil region mediating homodimerization.
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family. I-kappa-B kinase subfamily.
Research Fields
· Environmental Information Processing > Signal transduction > Ras signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Viral > Hepatitis C.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Measles.
· Human Diseases > Infectious diseases: Viral > Influenza A.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > Herpes simplex infection.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Organismal Systems > Immune system > Toll-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > RIG-I-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Cytosolic DNA-sensing pathway. (View pathway)
· Organismal Systems > Immune system > IL-17 signaling pathway. (View pathway)
References
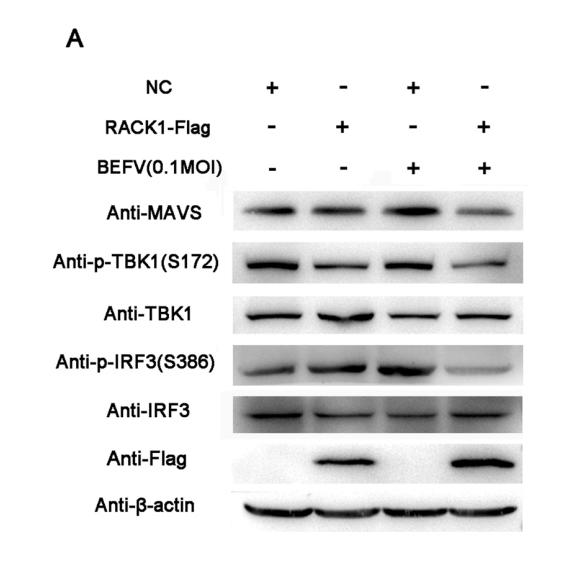
Application: WB Species: Human Sample: THP1 cells
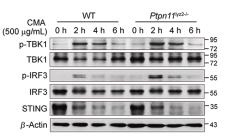
Application: WB Species: Mouse Sample: 3T3-L1 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.