Cyclin D1 Antibody - #DF6386
| Product: | Cyclin D1 Antibody |
| Catalog: | DF6386 |
| Description: | Rabbit polyclonal antibody to Cyclin D1 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Dog, Chicken, Xenopus |
| Mol.Wt.: | 34kDa; 34kD(Calculated). |
| Uniprot: | P24385 |
| RRID: | AB_2838349 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF6386, RRID:AB_2838349.
Fold/Unfold
AI327039;B cell CLL/lymphoma 1;B cell leukemia 1;B cell lymphoma 1 protein;B-cell lymphoma 1 protein;BCL 1;BCL-1;BCL-1 oncogene;BCL1;BCL1 oncogene;ccnd1;CCND1/FSTL3 fusion gene, included;CCND1/IGHG1 fusion gene, included;CCND1/IGLC1 fusion gene, included;CCND1/PTH fusion gene, included;CCND1_HUMAN;cD1;Cyl 1;D11S287E;G1/S specific cyclin D1;G1/S-specific cyclin-D1;Parathyroid adenomatosis 1;PRAD1;PRAD1 oncogene;U21B31;
Immunogens
A synthesized peptide derived from human Cyclin D1, corresponding to a region within C-terminal amino acids.
- P24385 CCND1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MEHQLLCCEVETIRRAYPDANLLNDRVLRAMLKAEETCAPSVSYFKCVQKEVLPSMRKIVATWMLEVCEEQKCEEEVFPLAMNYLDRFLSLEPVKKSRLQLLGATCMFVASKMKETIPLTAEKLCIYTDNSIRPEELLQMELLLVNKLKWNLAAMTPHDFIEHFLSKMPEAEENKQIIRKHAQTFVALCATDVKFISNPPSMVAAGSVVAAVQGLNLRSPNNFLSYYRLTRFLSRVIKCDPDCLRACQEQIEALLESSLRQAQQNMDPKAAEEEEEEEEEVDLACTPTDVRDVDI
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Regulatory component of the cyclin D1-CDK4 (DC) complex that phosphorylates and inhibits members of the retinoblastoma (RB) protein family including RB1 and regulates the cell-cycle during G(1)/S transition. Phosphorylation of RB1 allows dissociation of the transcription factor E2F from the RB/E2F complex and the subsequent transcription of E2F target genes which are responsible for the progression through the G(1) phase. Hypophosphorylates RB1 in early G(1) phase. Cyclin D-CDK4 complexes are major integrators of various mitogenenic and antimitogenic signals. Also substrate for SMAD3, phosphorylating SMAD3 in a cell-cycle-dependent manner and repressing its transcriptional activity. Component of the ternary complex, cyclin D1/CDK4/CDKN1B, required for nuclear translocation and activity of the cyclin D-CDK4 complex. Exhibits transcriptional corepressor activity with INSM1 on the NEUROD1 and INS promoters in a cell cycle-independent manner.
Phosphorylation at Thr-286 by MAP kinases is required for ubiquitination and degradation following DNA damage. It probably plays an essential role for recognition by the FBXO31 component of SCF (SKP1-cullin-F-box) protein ligase complex.
Ubiquitinated, primarily as 'Lys-48'-linked polyubiquitination. Ubiquitinated by a SCF (SKP1-CUL1-F-box protein) ubiquitin-protein ligase complex containing FBXO4 and CRYAB. Following DNA damage it is ubiquitinated by some SCF (SKP1-cullin-F-box) protein ligase complex containing FBXO31. SCF-type ubiquitination is dependent on Thr-286 phosphorylation (By similarity). Ubiquitinated also by UHRF2 apparently in a phosphorylation-independent manner. Ubiquitination leads to its degradation and G1 arrest. Deubiquitinated by USP2; leading to its stabilization.
Nucleus. Cytoplasm. Nucleus membrane.
Note: Cyclin D-CDK4 complexes accumulate at the nuclear membrane and are then translocated to the nucleus through interaction with KIP/CIP family members.
Belongs to the cyclin family. Cyclin D subfamily.
Research Fields
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Cellular Processes > Cell growth and death > p53 signaling pathway. (View pathway)
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Focal adhesion. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Tight junction. (View pathway)
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > AMPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Wnt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hedgehog signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Apelin signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hippo signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Jak-STAT signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > Endocrine resistance.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Measles.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Colorectal cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Pancreatic cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Endometrial cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Glioma. (View pathway)
· Human Diseases > Cancers: Specific types > Prostate cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Thyroid cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Melanoma. (View pathway)
· Human Diseases > Cancers: Specific types > Bladder cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Chronic myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Acute myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Small cell lung cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Non-small cell lung cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Breast cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Hepatocellular carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Gastric cancer. (View pathway)
· Human Diseases > Cardiovascular diseases > Viral myocarditis.
· Organismal Systems > Endocrine system > Prolactin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Thyroid hormone signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Oxytocin signaling pathway.
References
Application: WB Species: Mouse Sample:
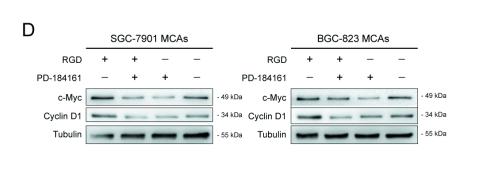
Application: WB Species: mouse Sample: BGC823MCAs and SGC7901MCAs
Application: WB Species: human Sample: Huh-7 cells
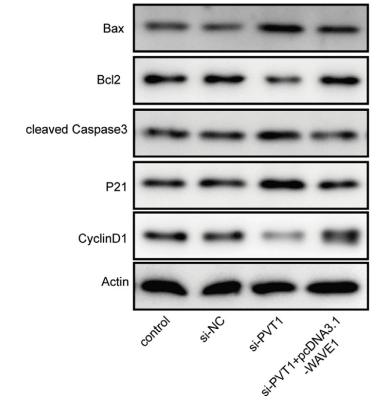
Application: WB Species: human Sample: ESCC cells
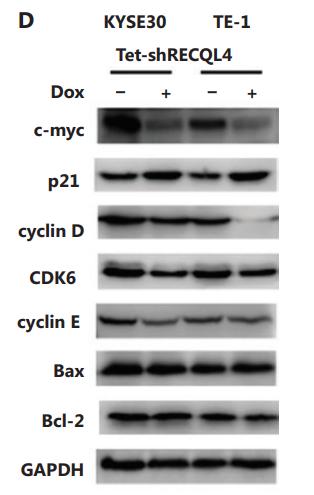
Application: WB Species: Human Sample: KYSE30 and TE-1 cells
Application: WB Species: Human Sample: tumour tissues
Application: WB Species: Mice Sample: tumor tissues
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.