ENO1 Antibody - #DF6191
| Product: | ENO1 Antibody |
| Catalog: | DF6191 |
| Description: | Rabbit polyclonal antibody to ENO1 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IHC |
| Reactivity: | Human, Mouse, Rat, Monkey |
| Prediction: | Pig, Bovine, Horse, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 47kDa; 47kD(Calculated). |
| Uniprot: | P06733 |
| RRID: | AB_2838157 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF6191, RRID:AB_2838157.
Fold/Unfold
2 phospho D glycerate hydro lyase; 2-phospho-D-glycerate hydro-lyase; Alpha enolase; Alpha enolase like 1; Alpha-enolase; C myc promoter binding protein; C-myc promoter-binding protein; EC 4.2.1.11; eno1; ENO1L1; ENOA_HUMAN; Enolase 1 (alpha); Enolase 1 (alpha) like 1; Enolase 1; Enolase alpha; MBP 1; MBP-1; MBP1; MBPB1; MPB 1; MPB-1; MPB1; MYC promoter binding protein 1; NNE; Non neural enolase; Non-neural enolase; Phosphopyruvate hydratase; Plasminogen binding protein; Plasminogen-binding protein; PPH; Tau crystallin;
Immunogens
A synthesized peptide derived from human ENO1, corresponding to a region within C-terminal amino acids.
The alpha/alpha homodimer is expressed in embryo and in most adult tissues. The alpha/beta heterodimer and the beta/beta homodimer are found in striated muscle, and the alpha/gamma heterodimer and the gamma/gamma homodimer in neurons.
- P06733 ENOA_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSILKIHAREIFDSRGNPTVEVDLFTSKGLFRAAVPSGASTGIYEALELRDNDKTRYMGKGVSKAVEHINKTIAPALVSKKLNVTEQEKIDKLMIEMDGTENKSKFGANAILGVSLAVCKAGAVEKGVPLYRHIADLAGNSEVILPVPAFNVINGGSHAGNKLAMQEFMILPVGAANFREAMRIGAEVYHNLKNVIKEKYGKDATNVGDEGGFAPNILENKEGLELLKTAIGKAGYTDKVVIGMDVAASEFFRSGKYDLDFKSPDDPSRYISPDQLADLYKSFIKDYPVVSIEDPFDQDDWGAWQKFTASAGIQVVGDDLTVTNPKRIAKAVNEKSCNCLLLKVNQIGSVTESLQACKLAQANGWGVMVSHRSGETEDTFIADLVVGLCTGQIKTGAPCRSERLAKYNQLLRIEEELGSKAKFAGRNFRNPLAK
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Glycolytic enzyme the catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate. In addition to glycolysis, involved in various processes such as growth control, hypoxia tolerance and allergic responses. May also function in the intravascular and pericellular fibrinolytic system due to its ability to serve as a receptor and activator of plasminogen on the cell surface of several cell-types such as leukocytes and neurons. Stimulates immunoglobulin production.
MBP1 binds to the myc promoter and acts as a transcriptional repressor. May be a tumor suppressor.
ISGylated.
Lysine 2-hydroxyisobutyrylation (Khib) by p300/EP300 activates the phosphopyruvate hydratase activity.
Cytoplasm. Cell membrane. Cytoplasm>Myofibril>Sarcomere>M line.
Note: Can translocate to the plasma membrane in either the homodimeric (alpha/alpha) or heterodimeric (alpha/gamma) form. ENO1 is localized to the M line.
Nucleus.
The alpha/alpha homodimer is expressed in embryo and in most adult tissues. The alpha/beta heterodimer and the beta/beta homodimer are found in striated muscle, and the alpha/gamma heterodimer and the gamma/gamma homodimer in neurons.
Belongs to the enolase family.
Research Fields
· Environmental Information Processing > Signal transduction > HIF-1 signaling pathway. (View pathway)
· Genetic Information Processing > Folding, sorting and degradation > RNA degradation.
· Metabolism > Carbohydrate metabolism > Glycolysis / Gluconeogenesis.
· Metabolism > Global and overview maps > Metabolic pathways.
· Metabolism > Global and overview maps > Carbon metabolism.
· Metabolism > Global and overview maps > Biosynthesis of amino acids.
References
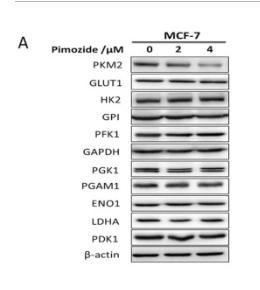
Application: WB Species: Human Sample: MCF-7 cells
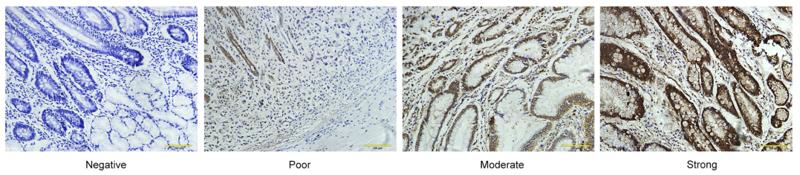
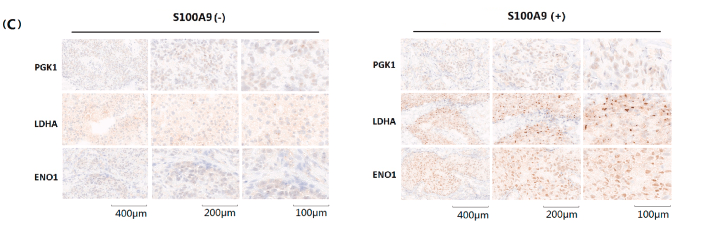
Application: IHC Species: human Sample: GC and adjacent normal tissues
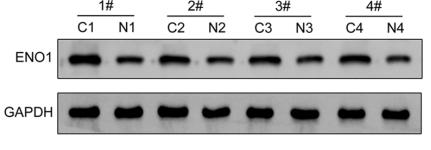
Application: WB Species: human Sample: GC tissue
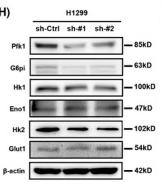
Application: WB Species: Human Sample: H1299 cells
Application: IHC Species: Human Sample: HER2+ BRCA tissues
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.