CDK4 Antibody - #DF6102
| Product: | CDK4 Antibody |
| Catalog: | DF6102 |
| Description: | Rabbit polyclonal antibody to CDK4 |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Xenopus |
| Mol.Wt.: | 34kDa; 34kD(Calculated). |
| Uniprot: | P11802 |
| RRID: | AB_2838070 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF6102, RRID:AB_2838070.
Fold/Unfold
Cdk 4; cdk4; CDK4 protein; CDK4_HUMAN; Cell division kinase 4; Cell division protein kinase 4; CMM 3; CMM3; Crk3; Cyclin dependent kinase 4; Cyclin-dependent kinase 4; Melanoma cutaneous malignant 3; MGC14458; p34 cdk4; PSK J3; PSK-J3;
Immunogens
A synthesized peptide derived from human CDK4, corresponding to a region within the internal amino acids.
- P11802 CDK4_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MATSRYEPVAEIGVGAYGTVYKARDPHSGHFVALKSVRVPNGGGGGGGLPISTVREVALLRRLEAFEHPNVVRLMDVCATSRTDREIKVTLVFEHVDQDLRTYLDKAPPPGLPAETIKDLMRQFLRGLDFLHANCIVHRDLKPENILVTSGGTVKLADFGLARIYSYQMALTPVVVTLWYRAPEVLLQSTYATPVDMWSVGCIFAEMFRRKPLFCGNSEADQLGKIFDLIGLPPEDDWPRDVSLPRGAFPPRGPRPVQSVVPEMEESGAQLLLEMLTFNPHKRISAFRALQHSYLHKDEGNPE
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Ser/Thr-kinase component of cyclin D-CDK4 (DC) complexes that phosphorylate and inhibit members of the retinoblastoma (RB) protein family including RB1 and regulate the cell-cycle during G(1)/S transition. Phosphorylation of RB1 allows dissociation of the transcription factor E2F from the RB/E2F complexes and the subsequent transcription of E2F target genes which are responsible for the progression through the G(1) phase. Hypophosphorylates RB1 in early G(1) phase. Cyclin D-CDK4 complexes are major integrators of various mitogenenic and antimitogenic signals. Also phosphorylates SMAD3 in a cell-cycle-dependent manner and represses its transcriptional activity. Component of the ternary complex, cyclin D/CDK4/CDKN1B, required for nuclear translocation and activity of the cyclin D-CDK4 complex.
Phosphorylation at Thr-172 is required for enzymatic activity. Phosphorylated, in vitro, at this site by CCNH-CDK7, but, in vivo, appears to be phosphorylated by a proline-directed kinase. In the cyclin D-CDK4-CDKN1B complex, this phosphorylation and consequent CDK4 enzyme activity, is dependent on the tyrosine phosphorylation state of CDKN1B. Thus, in proliferating cells, CDK4 within the complex is phosphorylated on Thr-172 in the T-loop. In resting cells, phosphorylation on Thr-172 is prevented by the non-tyrosine-phosphorylated form of CDKN1B.
Cytoplasm. Nucleus. Nucleus membrane.
Note: Cytoplasmic when non-complexed. Forms a cyclin D-CDK4 complex in the cytoplasm as cells progress through G(1) phase. The complex accumulates on the nuclear membrane and enters the nucleus on transition from G(1) to S phase. Also present in nucleoli and heterochromatin lumps. Colocalizes with RB1 after release into the nucleus.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. CDC2/CDKX subfamily.
Research Fields
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Cellular Processes > Cell growth and death > p53 signaling pathway. (View pathway)
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Tight junction. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > Endocrine resistance.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Measles.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Human Diseases > Cancers: Specific types > Pancreatic cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Glioma. (View pathway)
· Human Diseases > Cancers: Specific types > Melanoma. (View pathway)
· Human Diseases > Cancers: Specific types > Bladder cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Chronic myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Small cell lung cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Non-small cell lung cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Breast cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Hepatocellular carcinoma. (View pathway)
· Organismal Systems > Immune system > T cell receptor signaling pathway. (View pathway)
References
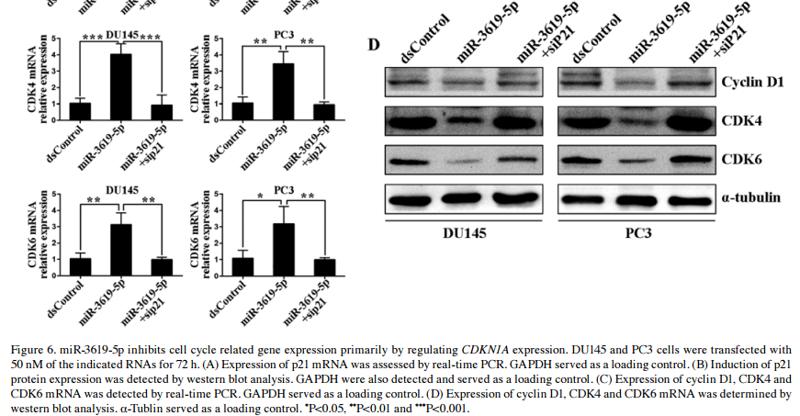
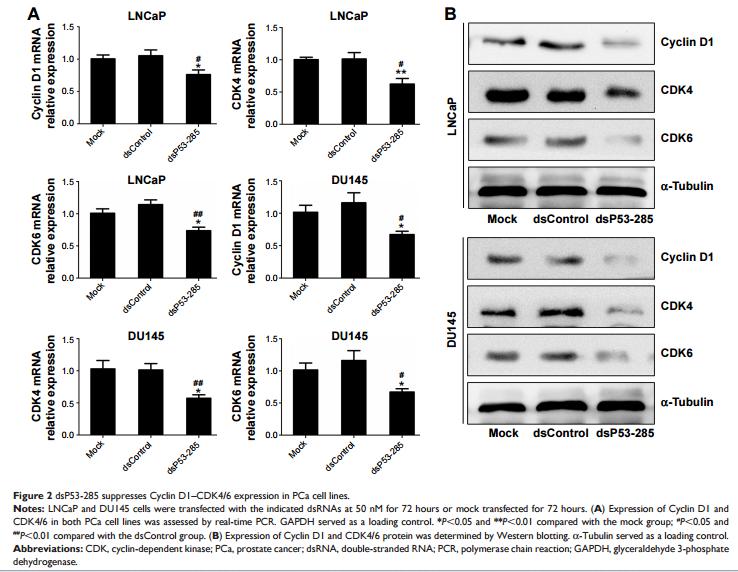
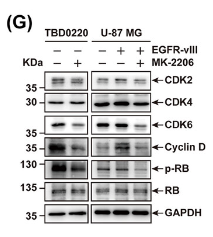
Application: WB Species: Human Sample: TBD0220, U-87 MG, and U-87 MG-EGFR-vIII cells
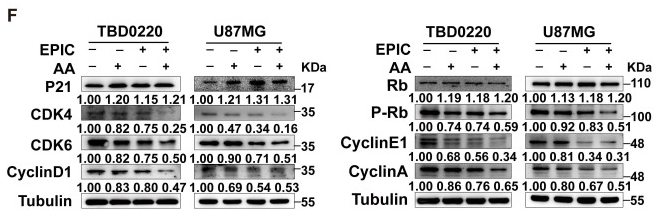
Application: WB Species: Human Sample: TBD0220 and U87MG cells
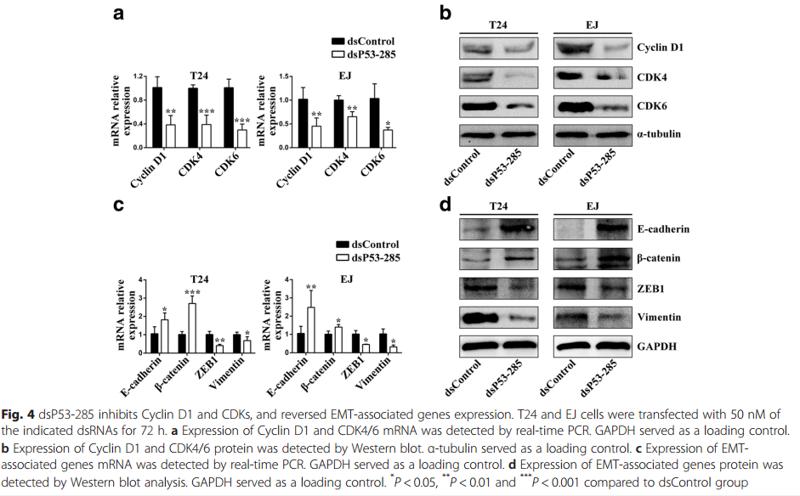
Application: WB Species: human Sample: T24 and EJ cells
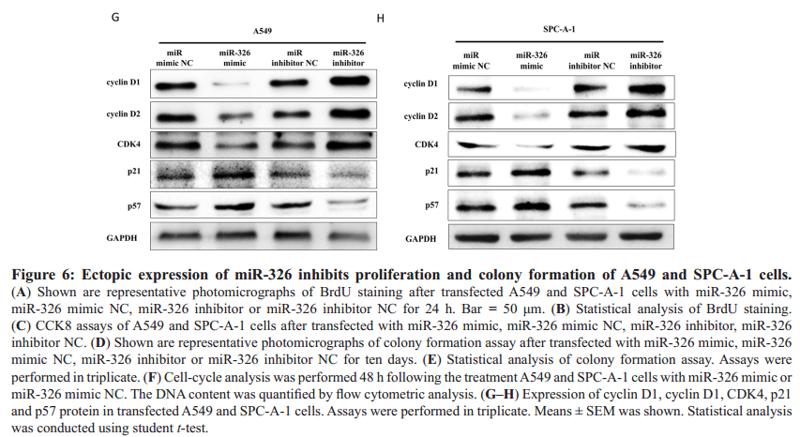
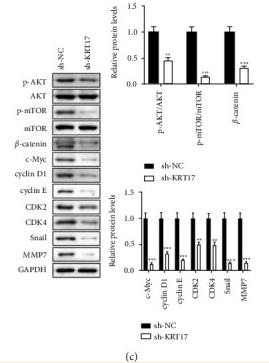
Application: WB Species: human Sample: BLCA cells
Application: WB Species: Rat Sample: testicular tissue
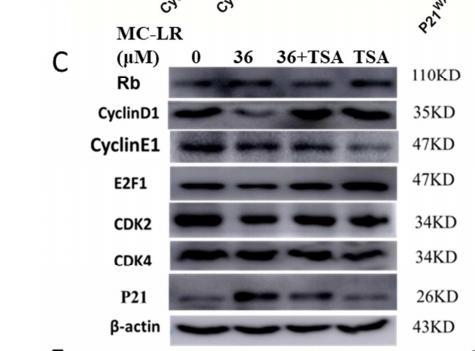
Application: WB Species: human Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.