ATF4 Antibody - #AF5416
| Product: | ATF4 Antibody |
| Catalog: | AF5416 |
| Description: | Rabbit polyclonal antibody to ATF4 |
| Application: | WB |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse |
| Prediction: | Pig, Bovine, Dog |
| Mol.Wt.: | 39~49kD; 39kD(Calculated). |
| Uniprot: | P18848 |
| RRID: | AB_2837900 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF5416, RRID:AB_2837900.
Fold/Unfold
Activating transcription factor 4; ATF 4; ATF4; ATF4 protein; ATF4_HUMAN; cAMP-dependent transcription factor ATF-4; cAMP-responsive element-binding protein 2; CREB 2; CREB-2; CREB2; Cyclic AMP dependent transcription factor ATF 4; Cyclic AMP response element binding protein 2; Cyclic AMP-dependent transcription factor ATF-4; Cyclic AMP-responsive element-binding protein 2; DNA binding protein TAXREB67; DNA-binding protein TAXREB67; Tax Responsive Enhancer Element B67; Tax-responsive enhancer element-binding protein 67; TaxREB67; TXREB;
Immunogens
A synthesized peptide derived from human ATF4, corresponding to a region within the internal amino acids.
- P18848 ATF4_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MTEMSFLSSEVLVGDLMSPFDQSGLGAEESLGLLDDYLEVAKHFKPHGFSSDKAKAGSSEWLAVDGLVSPSNNSKEDAFSGTDWMLEKMDLKEFDLDALLGIDDLETMPDDLLTTLDDTCDLFAPLVQETNKQPPQTVNPIGHLPESLTKPDQVAPFTFLQPLPLSPGVLSSTPDHSFSLELGSEVDITEGDRKPDYTAYVAMIPQCIKEEDTPSDNDSGICMSPESYLGSPQHSPSTRGSPNRSLPSPGVLCGSARPKPYDPPGEKMVAAKVKGEKLDKKLKKMEQNKTAATRYRQKKRAEQEALTGECKELEKKNEALKERADSLAKEIQYLKDLIEEVRKARGKKRVP
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Transcriptional activator. Binds the cAMP response element (CRE) (consensus: 5'-GTGACGT[AC][AG]-3'), a sequence present in many viral and cellular promoters. Cooperates with FOXO1 in osteoblasts to regulate glucose homeostasis through suppression of beta-cell production and decrease in insulin production (By similarity). It binds to a Tax-responsive enhancer element in the long terminal repeat of HTLV-I. Regulates the induction of DDIT3/CHOP and asparagine synthetase (ASNS) in response to endoplasmic reticulum (ER) stress. In concert with DDIT3/CHOP, activates the transcription of TRIB3 and promotes ER stress-induced neuronal apoptosis by regulating the transcriptional induction of BBC3/PUMA. Activates transcription of SIRT4. Regulates the circadian expression of the core clock component PER2 and the serotonin transporter SLC6A4. Binds in a circadian time-dependent manner to the cAMP response elements (CRE) in the SLC6A4 and PER2 promoters and periodically activates the transcription of these genes. During ER stress response, activates the transcription of NLRP1, possibly in concert with other factors.
Ubiquitinated by SCF(BTRC) in response to mTORC1 signal, followed by proteasomal degradation and leading to down-regulate expression of SIRT4.
Phosphorylated by NEK6 (By similarity). Phosphorylated on the betaTrCP degron motif at Ser-219, followed by phosphorylation at Thr-213, Ser-224, Ser-231, Ser-235 and Ser-248, promoting interaction with BTRC and ubiquitination. Phosphorylation is promoted by mTORC1 (By similarity).
Phosphorylated by NEK6.
Cytoplasm. Cell membrane. Nucleus. Cytoplasm>Cytoskeleton>Microtubule organizing center>Centrosome.
Note: Colocalizes with GABBR1 in hippocampal neuron dendritic membranes (By similarity). Colocalizes with NEK6 at the centrosome (PubMed:20873783).
The BetaTrCP degron motif promotes binding to BTRC when phosphorylated.
Belongs to the bZIP family.
Research Fields
· Cellular Processes > Cell growth and death > Apoptosis. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cGMP-PKG signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > TNF signaling pathway. (View pathway)
· Genetic Information Processing > Folding, sorting and degradation > Protein processing in endoplasmic reticulum. (View pathway)
· Human Diseases > Endocrine and metabolic diseases > Non-alcoholic fatty liver disease (NAFLD).
· Human Diseases > Substance dependence > Cocaine addiction.
· Human Diseases > Substance dependence > Amphetamine addiction.
· Human Diseases > Substance dependence > Alcoholism.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Human Diseases > Cancers: Specific types > Prostate cancer. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway. (View pathway)
· Organismal Systems > Circulatory system > Adrenergic signaling in cardiomyocytes. (View pathway)
· Organismal Systems > Nervous system > Long-term potentiation.
· Organismal Systems > Nervous system > Neurotrophin signaling pathway. (View pathway)
· Organismal Systems > Nervous system > Cholinergic synapse.
· Organismal Systems > Nervous system > Dopaminergic synapse.
· Organismal Systems > Endocrine system > Insulin secretion. (View pathway)
· Organismal Systems > Endocrine system > Estrogen signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Thyroid hormone synthesis.
· Organismal Systems > Endocrine system > Glucagon signaling pathway.
· Organismal Systems > Endocrine system > Aldosterone synthesis and secretion.
· Organismal Systems > Endocrine system > Relaxin signaling pathway.
References
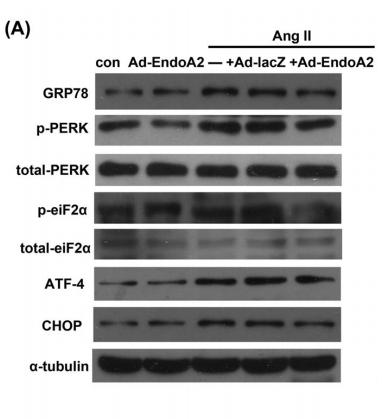
Application: WB Species: rat Sample: renal
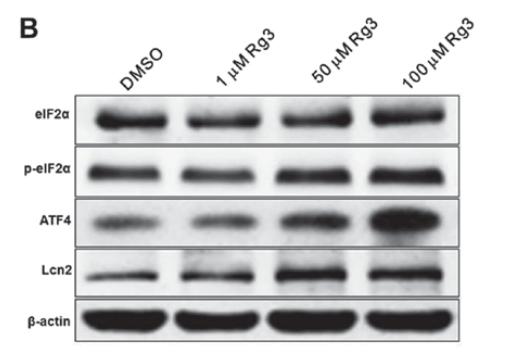
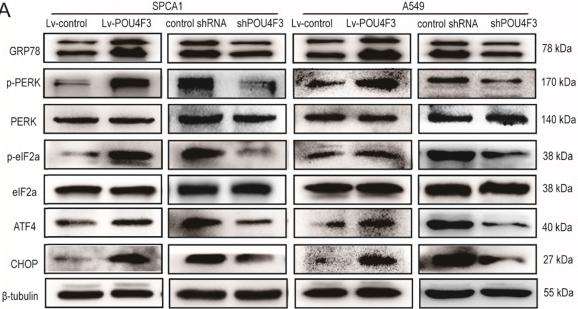
Application: WB Species: Human Sample: LUAD cells
Application: WB Species: rat Sample: cardiomyocytes
Application: WB Species: human Sample: GBC‑SD cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.