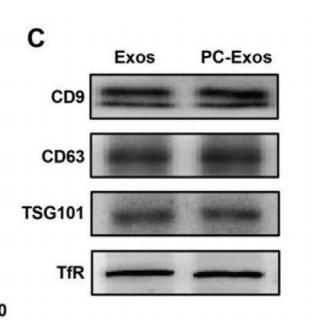
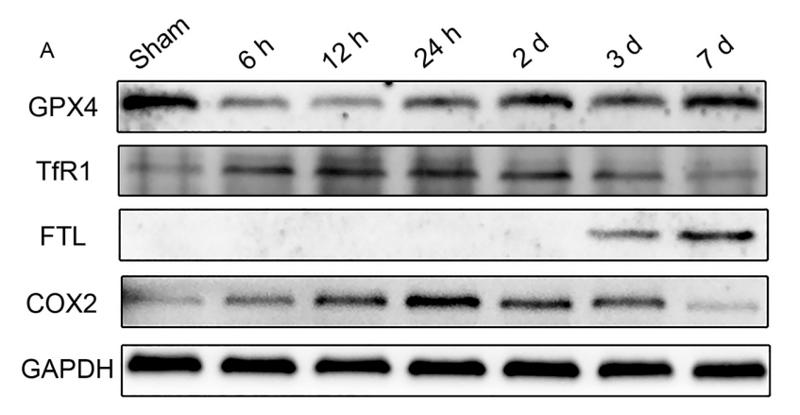
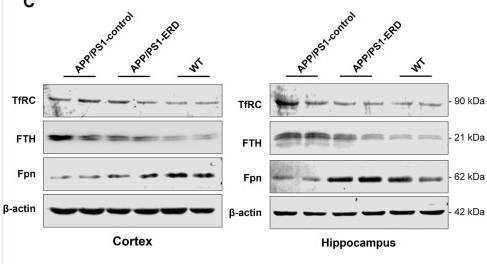
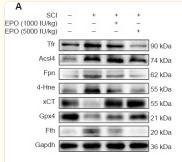
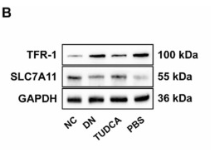
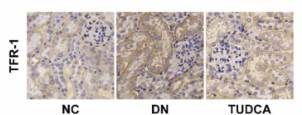
Transferrin Receptor Antibody - #AF5343
| Product: | Transferrin Receptor Antibody |
| Catalog: | AF5343 |
| Description: | Rabbit polyclonal antibody to Transferrin Receptor |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IHC, IF/ICC |
| Reactivity: | Human, Mouse, Monkey |
| Prediction: | Rat, Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken |
| Mol.Wt.: | 50kD(reduced),80~130kDa; 85kD(Calculated). |
| Uniprot: | P02786 |
| RRID: | AB_2837828 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF5343, RRID:AB_2837828.
Fold/Unfold
CD 71; CD71; CD71 antigen; IMD46; OTTHUMP00000208523; OTTHUMP00000208524; OTTHUMP00000208525; p90; sTfR; T9; TFR 1; TfR; TfR1; TFR1_HUMAN; TFRC; TR; Transferrin receptor (p90 CD71); Transferrin receptor protein 1, serum form; Trfr;
Immunogens
A synthesized peptide derived from human Transferrin Receptor
- P02786 TFR1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MMDQARSAFSNLFGGEPLSYTRFSLARQVDGDNSHVEMKLAVDEEENADNNTKANVTKPKRCSGSICYGTIAVIVFFLIGFMIGYLGYCKGVEPKTECERLAGTESPVREEPGEDFPAARRLYWDDLKRKLSEKLDSTDFTGTIKLLNENSYVPREAGSQKDENLALYVENQFREFKLSKVWRDQHFVKIQVKDSAQNSVIIVDKNGRLVYLVENPGGYVAYSKAATVTGKLVHANFGTKKDFEDLYTPVNGSIVIVRAGKITFAEKVANAESLNAIGVLIYMDQTKFPIVNAELSFFGHAHLGTGDPYTPGFPSFNHTQFPPSRSSGLPNIPVQTISRAAAEKLFGNMEGDCPSDWKTDSTCRMVTSESKNVKLTVSNVLKEIKILNIFGVIKGFVEPDHYVVVGAQRDAWGPGAAKSGVGTALLLKLAQMFSDMVLKDGFQPSRSIIFASWSAGDFGSVGATEWLEGYLSSLHLKAFTYINLDKAVLGTSNFKVSASPLLYTLIEKTMQNVKHPVTGQFLYQDSNWASKVEKLTLDNAAFPFLAYSGIPAVSFCFCEDTDYPYLGTTMDTYKELIERIPELNKVARAAAEVAGQFVIKLTHDVELNLDYERYNSQLLSFVRDLNQYRADIKEMGLSLQWLYSARGDFFRATSRLTTDFGNAEKTDRFVMKKLNDRVMRVEYHFLSPYVSPKESPFRHVFWGSGSHTLPALLENLKLRKQNNGAFNETLFRNQLALATWTIQGAANALSGDVWDIDNEF
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Cellular uptake of iron occurs via receptor-mediated endocytosis of ligand-occupied transferrin receptor into specialized endosomes. Endosomal acidification leads to iron release. The apotransferrin-receptor complex is then recycled to the cell surface with a return to neutral pH and the concomitant loss of affinity of apotransferrin for its receptor. Transferrin receptor is necessary for development of erythrocytes and the nervous system (By similarity). A second ligand, the heditary hemochromatosis protein HFE, competes for binding with transferrin for an overlapping C-terminal binding site. Positively regulates T and B cell proliferation through iron uptake.
(Microbial infection) Acts as a receptor for new-world arenaviruses: Guanarito, Junin and Machupo virus.
N- and O-glycosylated, phosphorylated and palmitoylated. The serum form is only glycosylated.
Proteolytically cleaved on Arg-100 to produce the soluble serum form (sTfR).
Palmitoylated on both Cys-62 and Cys-67. Cys-62 seems to be the major site of palmitoylation.
Cell membrane>Single-pass type II membrane protein. Melanosome.
Note: Identified by mass spectrometry in melanosome fractions from stage I to stage IV.
Secreted.
Belongs to the peptidase M28 family. M28B subfamily.
Research Fields
· Cellular Processes > Transport and catabolism > Endocytosis. (View pathway)
· Cellular Processes > Transport and catabolism > Phagosome. (View pathway)
· Cellular Processes > Cell growth and death > Ferroptosis. (View pathway)
· Environmental Information Processing > Signal transduction > HIF-1 signaling pathway. (View pathway)
· Organismal Systems > Immune system > Hematopoietic cell lineage. (View pathway)
References
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.