RASH/RASK/RASN Antibody - #AF0247
| Product: | RASH/RASK/RASN Antibody |
| Catalog: | AF0247 |
| Description: | Rabbit polyclonal antibody to RASH/RASK/RASN |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 25kDa,21kDa; 21kD,22kD(Calculated). |
| Uniprot: | P01111 | P01112 | P01116 |
| RRID: | AB_2833422 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0247, RRID:AB_2833422.
Fold/Unfold
ALPS4; AV095280; GTPase NRas; HRAS1; N ras; N ras protein part 4; Neuroblastoma RAS viral (v ras) oncogene homolog; NRAS; NRAS1; NS6; OTTHUMP00000013879; OTTMUSP00000023521; RASN_HUMAN; Transforming protein N Ras; Transforming protein N-Ras; v ras neuroblastoma RAS viral oncogene homolog; C BAS/HAS; c H ras; C HA RAS1; c has/bas p21 protein; c ras Ki 2 activated oncogene; c-H-ras; CTLO; GTP and GDP binding peptide B; GTPase HRas, N-terminally processed; H Ras 1; H RASIDX; H-Ras-1; Ha Ras; Ha Ras1 proto oncoprotein; Ha-Ras; HAMSV; Harvey rat sarcoma viral oncogene homolog; Harvey rat sarcoma viral oncoprotein; HRAS; HRAS1; K ras; N ras; p19 H RasIDX protein; p21ras; Ras family small GTP binding protein H Ras; RASH_HUMAN; RASH1; Transformation gene oncogene HAMSV; Transforming protein p21; v Ha ras Harvey rat sarcoma viral oncogene homolog; VH Ras; vHa RAS; c Ki ras2; c Kirsten ras protein; c-K-ras; c-Ki-ras; Cellular c Ki ras2 proto oncogene; Cellular transforming proto oncogene; CFC2; cK Ras; GTPase KRas; K RAS p21 protein; K RAS2A; K RAS2B; K RAS4A; K RAS4B; K-Ras 2; KI RAS; Ki-Ras; KIRSTEN MURINE SARCOMA VIRUS 2; Kirsten rat sarcoma 2 viral (v Ki ras2) oncogene homolog; Kirsten rat sarcoma viral oncogene homolog; KRAS; KRAS proto oncogene, GTPase; KRAS1; KRAS2; N-terminally processed; NS; NS3; Oncogene KRAS2; p21ras; PR310 c K ras oncogene; PR310 cK ras oncogene; RALD; RASK_HUMAN; RASK2; Transforming protein p21; v Ki ras2 Kirsten rat sarcoma 2 viral oncogene homolog; v Ki ras2 Kirsten rat sarcoma viral oncogene homolog;
Immunogens
A synthesized peptide derived from human RASH/RASK/RASN, corresponding to a region within N-terminal amino acids.
- P01111 RASN_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQVVIDGETCLLDILDTAGQEEYSAMRDQYMRTGEGFLCVFAINNSKSFADINLYREQIKRVKDSDDVPMVLVGNKCDLPTRTVDTKQAHELAKSYGIPFIETSAKTRQGVEDAFYTLVREIRQYRMKKLNSSDDGTQGCMGLPCVVM
- P01112 RASH_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQVVIDGETCLLDILDTAGQEEYSAMRDQYMRTGEGFLCVFAINNTKSFEDIHQYREQIKRVKDSDDVPMVLVGNKCDLAARTVESRQAQDLARSYGIPYIETSAKTRQGVEDAFYTLVREIRQHKLRKLNPPDESGPGCMSCKCVLS
- P01116 RASK_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQVVIDGETCLLDILDTAGQEEYSAMRDQYMRTGEGFLCVFAINNTKSFEDIHHYREQIKRVKDSEDVPMVLVGNKCDLPSRTVDTKQAQDLARSYGIPFIETSAKTRQRVEDAFYTLVREIRQYRLKKISKEEKTPGCVKIKKCIIM
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Ras proteins bind GDP/GTP and possess intrinsic GTPase activity.
Palmitoylated by the ZDHHC9-GOLGA7 complex. Depalmitoylated by ABHD17A, ABHD17B and ABHD17C. A continuous cycle of de- and re-palmitoylation regulates rapid exchange between plasma membrane and Golgi.
Acetylation at Lys-104 prevents interaction with guanine nucleotide exchange factors (GEFs).
Ubiquitinated by the BCR(LZTR1) E3 ubiquitin ligase complex at Lys-170 in a non-degradative manner, leading to inhibit Ras signaling by decreasing Ras association with membranes.
Phosphorylation at Ser-89 by STK19 enhances NRAS-association with its downstream effectors.
Cell membrane>Lipid-anchor>Cytoplasmic side. Golgi apparatus membrane>Lipid-anchor.
Note: Shuttles between the plasma membrane and the Golgi apparatus.
Belongs to the small GTPase superfamily. Ras family.
Involved in the activation of Ras protein signal transduction. Ras proteins bind GDP/GTP and possess intrinsic GTPase activity.
Palmitoylated by the ZDHHC9-GOLGA7 complex. A continuous cycle of de- and re-palmitoylation regulates rapid exchange between plasma membrane and Golgi.
S-nitrosylated; critical for redox regulation. Important for stimulating guanine nucleotide exchange. No structural perturbation on nitrosylation.
The covalent modification of cysteine by 15-deoxy-Delta12,14-prostaglandin-J2 is autocatalytic and reversible. It may occur as an alternative to other cysteine modifications, such as S-nitrosylation and S-palmitoylation.
Acetylation at Lys-104 prevents interaction with guanine nucleotide exchange factors (GEFs).
Ubiquitinated by the BCR(LZTR1) E3 ubiquitin ligase complex at Lys-170 in a non-degradative manner, leading to inhibit Ras signaling by decreasing Ras association with membranes.
Cell membrane>Lipid-anchor>Cytoplasmic side. Golgi apparatus. Golgi apparatus membrane>Lipid-anchor.
Note: The active GTP-bound form is localized most strongly to membranes than the inactive GDP-bound form (By similarity). Shuttles between the plasma membrane and the Golgi apparatus.
Nucleus. Cytoplasm. Cytoplasm>Perinuclear region.
Note: Colocalizes with RACK1 to the perinuclear region.
Widely expressed.
Belongs to the small GTPase superfamily. Ras family.
Ras proteins bind GDP/GTP and possess intrinsic GTPase activity. Plays an important role in the regulation of cell proliferation. Plays a role in promoting oncogenic events by inducing transcriptional silencing of tumor suppressor genes (TSGs) in colorectal cancer (CRC) cells in a ZNF304-dependent manner.
Acetylation at Lys-104 prevents interaction with guanine nucleotide exchange factors (GEFs).
Ubiquitinated by the BCR(LZTR1) E3 ubiquitin ligase complex at Lys-170 in a non-degradative manner, leading to inhibit Ras signaling by decreasing Ras association with membranes.
Cell membrane>Lipid-anchor>Cytoplasmic side. Cytoplasm>Cytosol.
Belongs to the small GTPase superfamily. Ras family.
Research Fields
· Cellular Processes > Transport and catabolism > Autophagy - animal. (View pathway)
· Cellular Processes > Transport and catabolism > Endocytosis. (View pathway)
· Cellular Processes > Cell growth and death > Apoptosis. (View pathway)
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Focal adhesion. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Gap junction. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Signaling pathways regulating pluripotency of stem cells. (View pathway)
· Cellular Processes > Cell motility > Regulation of actin cytoskeleton. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > ErbB signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Ras signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Rap1 signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Sphingolipid signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Phospholipase D signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > mTOR signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Apelin signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Jak-STAT signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > EGFR tyrosine kinase inhibitor resistance.
· Human Diseases > Drug resistance: Antineoplastic > Endocrine resistance.
· Human Diseases > Substance dependence > Alcoholism.
· Human Diseases > Infectious diseases: Viral > Hepatitis C.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Colorectal cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Renal cell carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Pancreatic cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Endometrial cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Glioma. (View pathway)
· Human Diseases > Cancers: Specific types > Prostate cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Thyroid cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Melanoma. (View pathway)
· Human Diseases > Cancers: Specific types > Bladder cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Chronic myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Acute myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Non-small cell lung cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Breast cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Hepatocellular carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Gastric cancer. (View pathway)
· Human Diseases > Cancers: Overview > Central carbon metabolism in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Choline metabolism in cancer. (View pathway)
· Organismal Systems > Immune system > Chemokine signaling pathway. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway - multiple species. (View pathway)
· Organismal Systems > Development > Axon guidance. (View pathway)
· Organismal Systems > Immune system > Natural killer cell mediated cytotoxicity. (View pathway)
· Organismal Systems > Immune system > T cell receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > B cell receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Fc epsilon RI signaling pathway. (View pathway)
· Organismal Systems > Nervous system > Long-term potentiation.
· Organismal Systems > Nervous system > Neurotrophin signaling pathway. (View pathway)
· Organismal Systems > Nervous system > Cholinergic synapse.
· Organismal Systems > Nervous system > Serotonergic synapse.
· Organismal Systems > Nervous system > Long-term depression.
· Organismal Systems > Endocrine system > Insulin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Progesterone-mediated oocyte maturation.
· Organismal Systems > Endocrine system > Estrogen signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Melanogenesis.
· Organismal Systems > Endocrine system > Prolactin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Thyroid hormone signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Oxytocin signaling pathway.
· Organismal Systems > Endocrine system > Relaxin signaling pathway.
· Organismal Systems > Excretory system > Aldosterone-regulated sodium reabsorption.
References
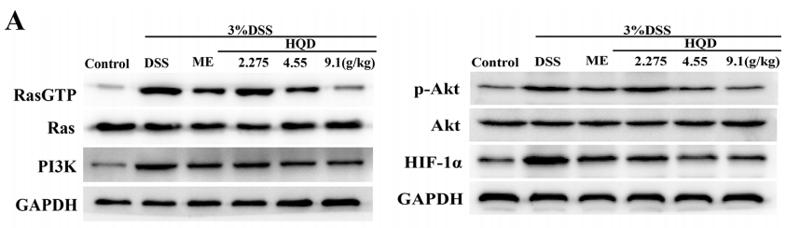
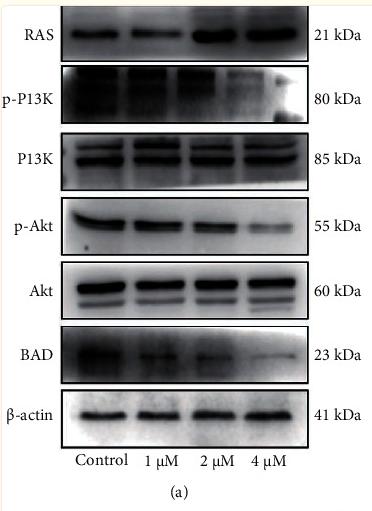
Application: WB Species: mouse Sample: colorectal
Application: WB Species: Human Sample: HSC-3 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.