HER2/ErbB2 Antibody - #AF7681
| Product: | HER2/ErbB2 Antibody |
| Catalog: | AF7681 |
| Description: | Rabbit polyclonal antibody to HER2/ErbB2 |
| Application: | WB IF/ICC |
| Cited expt.: | WB, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 138kDa; 138kD(Calculated). |
| Uniprot: | P04626 |
| RRID: | AB_2844045 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF7681, RRID:AB_2844045.
Fold/Unfold
Verb b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog; C erb B2/neu protein; CD340; CD340 antigen; Cerb B2/neu protein; CerbB2; Erb b2 receptor tyrosine kinase 2; ERBB2; ERBB2_HUMAN; HER 2; HER 2/NEU; HER2; Herstatin; Human epidermal growth factor receptor 2; Metastatic lymph node gene 19 protein; MLN 19; MLN19; NEU; NEU proto oncogene; Neuro/glioblastoma derived oncogene homolog; Neuroblastoma/glioblastoma derived oncogene homolog; NGL; p185erbB2; Proto-oncogene c-ErbB-2; Proto-oncogene Neu; Receptor tyrosine-protein kinase erbB-2; TKR1; Tyrosine kinase type cell surface receptor HER2; Tyrosine kinase-type cell surface receptor HER2; V erb b2 avian erythroblastic leukemia viral oncogene homolog 2 (neuro/glioblastoma derived oncogene homolog); V erb b2 avian erythroblastic leukemia viral oncogene homolog 2; V erb b2 avian erythroblastic leukemia viral oncoprotein 2; V erb b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian); V erb b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog; Verb b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian);
Immunogens
A synthesized peptide derived from human HER2/ErbB2, corresponding to a region within the internal amino acids.
Expressed in a variety of tumor tissues including primary breast tumors and tumors from small bowel, esophagus, kidney and mouth.
- P04626 ERBB2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MELAALCRWGLLLALLPPGAASTQVCTGTDMKLRLPASPETHLDMLRHLYQGCQVVQGNLELTYLPTNASLSFLQDIQEVQGYVLIAHNQVRQVPLQRLRIVRGTQLFEDNYALAVLDNGDPLNNTTPVTGASPGGLRELQLRSLTEILKGGVLIQRNPQLCYQDTILWKDIFHKNNQLALTLIDTNRSRACHPCSPMCKGSRCWGESSEDCQSLTRTVCAGGCARCKGPLPTDCCHEQCAAGCTGPKHSDCLACLHFNHSGICELHCPALVTYNTDTFESMPNPEGRYTFGASCVTACPYNYLSTDVGSCTLVCPLHNQEVTAEDGTQRCEKCSKPCARVCYGLGMEHLREVRAVTSANIQEFAGCKKIFGSLAFLPESFDGDPASNTAPLQPEQLQVFETLEEITGYLYISAWPDSLPDLSVFQNLQVIRGRILHNGAYSLTLQGLGISWLGLRSLRELGSGLALIHHNTHLCFVHTVPWDQLFRNPHQALLHTANRPEDECVGEGLACHQLCARGHCWGPGPTQCVNCSQFLRGQECVEECRVLQGLPREYVNARHCLPCHPECQPQNGSVTCFGPEADQCVACAHYKDPPFCVARCPSGVKPDLSYMPIWKFPDEEGACQPCPINCTHSCVDLDDKGCPAEQRASPLTSIISAVVGILLVVVLGVVFGILIKRRQQKIRKYTMRRLLQETELVEPLTPSGAMPNQAQMRILKETELRKVKVLGSGAFGTVYKGIWIPDGENVKIPVAIKVLRENTSPKANKEILDEAYVMAGVGSPYVSRLLGICLTSTVQLVTQLMPYGCLLDHVRENRGRLGSQDLLNWCMQIAKGMSYLEDVRLVHRDLAARNVLVKSPNHVKITDFGLARLLDIDETEYHADGGKVPIKWMALESILRRRFTHQSDVWSYGVTVWELMTFGAKPYDGIPAREIPDLLEKGERLPQPPICTIDVYMIMVKCWMIDSECRPRFRELVSEFSRMARDPQRFVVIQNEDLGPASPLDSTFYRSLLEDDDMGDLVDAEEYLVPQQGFFCPDPAPGAGGMVHHRHRSSSTRSGGGDLTLGLEPSEEEAPRSPLAPSEGAGSDVFDGDLGMGAAKGLQSLPTHDPSPLQRYSEDPTVPLPSETDGYVAPLTCSPQPEYVNQPDVRPQPPSPREGPLPAARPAGATLERPKTLSPGKNGVVKDVFAFGGAVENPEYLTPQGGAAPQPHPPPAFSPAFDNLYYWDQDPPERGAPPSTFKGTPTAENPEYLGLDVPV
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Protein tyrosine kinase that is part of several cell surface receptor complexes, but that apparently needs a coreceptor for ligand binding. Essential component of a neuregulin-receptor complex, although neuregulins do not interact with it alone. GP30 is a potential ligand for this receptor. Regulates outgrowth and stabilization of peripheral microtubules (MTs). Upon ERBB2 activation, the MEMO1-RHOA-DIAPH1 signaling pathway elicits the phosphorylation and thus the inhibition of GSK3B at cell membrane. This prevents the phosphorylation of APC and CLASP2, allowing its association with the cell membrane. In turn, membrane-bound APC allows the localization of MACF1 to the cell membrane, which is required for microtubule capture and stabilization.
In the nucleus is involved in transcriptional regulation. Associates with the 5'-TCAAATTC-3' sequence in the PTGS2/COX-2 promoter and activates its transcription. Implicated in transcriptional activation of CDKN1A; the function involves STAT3 and SRC. Involved in the transcription of rRNA genes by RNA Pol I and enhances protein synthesis and cell growth.
Autophosphorylated. Autophosphorylation occurs in trans, i.e. one subunit of the dimeric receptor phosphorylates tyrosine residues on the other subunit (Probable). Ligand-binding increases phosphorylation on tyrosine residues. Signaling via SEMA4C promotes phosphorylation at Tyr-1248. Dephosphorylated by PTPN12.
Cell membrane>Single-pass type I membrane protein. Early endosome. Cytoplasm>Perinuclear region. Nucleus.
Note: Translocation to the nucleus requires endocytosis, probably endosomal sorting and is mediated by importin beta-1/KPNB1. Also detected in VPS35-positive endosome-to-TGN retrograde vesicles (PubMed:31138794).
Cytoplasm. Nucleus.
Cytoplasm. Nucleus.
Expressed in a variety of tumor tissues including primary breast tumors and tumors from small bowel, esophagus, kidney and mouth.
Belongs to the protein kinase superfamily. Tyr protein kinase family. EGF receptor subfamily.
Research Fields
· Cellular Processes > Cellular community - eukaryotes > Focal adhesion. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Adherens junction. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Tight junction. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > ErbB signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Calcium signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > HIF-1 signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > EGFR tyrosine kinase inhibitor resistance.
· Human Diseases > Drug resistance: Antineoplastic > Endocrine resistance.
· Human Diseases > Drug resistance: Antineoplastic > Platinum drug resistance.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Pancreatic cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Endometrial cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Prostate cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Bladder cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Non-small cell lung cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Breast cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Gastric cancer. (View pathway)
· Human Diseases > Cancers: Overview > Central carbon metabolism in cancer. (View pathway)
References
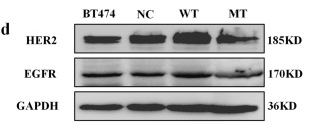
Application: WB Species: Human Sample: breast cancer
Application: IHC Species: Human Sample: BT474 cell
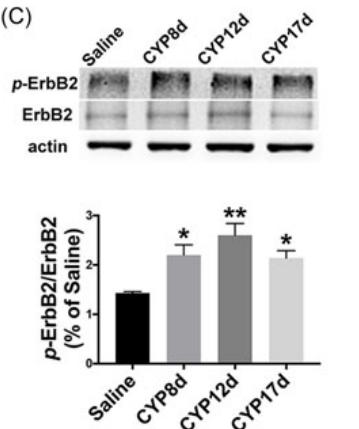
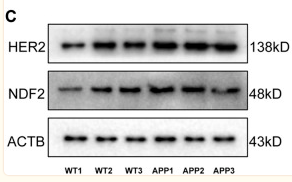
Application: WB Species: Mouse Sample: brain tissue
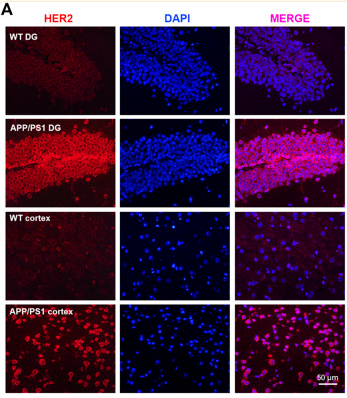
Application: IF/ICC Species: Mouse Sample: brain tissue
Application: WB Species: rat Sample: spinal dorsal horn
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.