HES1 Antibody - #AF7575
| Product: | HES1 Antibody |
| Catalog: | AF7575 |
| Description: | Rabbit polyclonal antibody to HES1 |
| Application: | WB |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Chicken, Xenopus |
| Mol.Wt.: | 30kDa; 30kD(Calculated). |
| Uniprot: | Q14469 |
| RRID: | AB_2843939 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF7575, RRID:AB_2843939.
Fold/Unfold
bHLHb39; C-HAIRY1; c-hairy1A; Class B basic helix-loop-helix protein 39; FLJ20408; Hairy and enhancer of split 1 (Drosophila); Hairy and enhancer of split 1; Hairy homolog (Drosophila); Hairy homolog; Hairy like; Hairy, Drosophila, homolog of; Hairy-like protein; Hairy/enhancer of split, Drosophila, homolog of, 1; HAIRY1; HES-1; hes1; Hes1 hairy and enhancer of split 1 (Drosophila); HES1_HUMAN; HHL; HL; HRY; MGC129109; OTTHUMP00000209031; RHL; Transcription factor HES-1;
Immunogens
A synthesized peptide derived from human HES1, corresponding to a region within N-terminal amino acids.
- Q14469 HES1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MPADIMEKNSSSPVAATPASVNTTPDKPKTASEHRKSSKPIMEKRRRARINESLSQLKTLILDALKKDSSRHSKLEKADILEMTVKHLRNLQRAQMTAALSTDPSVLGKYRAGFSECMNEVTRFLSTCEGVNTEVRTRLLGHLANCMTQINAMTYPGQPHPALQAPPPPPPGPGGPQHAPFAPPPPLVPIPGGAAPPPGGAPCKLGSQAGEAAKVFGGFQVVPAPDGQFAFLIPNGAFAHSGPVIPVYTSNSGTSVGPNAVSPSSGPSLTADSMWRPWRN
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Transcriptional repressor of genes that require a bHLH protein for their transcription. May act as a negative regulator of myogenesis by inhibiting the functions of MYOD1 and ASH1. Binds DNA on N-box motifs: 5'-CACNAG-3' with high affinity and on E-box motifs: 5'-CANNTG-3' with low affinity (By similarity). May play a role in a functional FA core complex response to DNA cross-link damage, being required for the stability and nuclear localization of FA core complex proteins, as well as for FANCD2 monoubiquitination in response to DNA damage.
Nucleus.
Has a particular type of basic domain (presence of a helix-interrupting proline) that binds to the N-box (CACNAG), rather than the canonical E-box (CANNTG).
The C-terminal WRPW motif is a transcriptional repression domain necessary for the interaction with Groucho/TLE family members, transcriptional corepressors recruited to specific target DNA by Hairy-related proteins.
The bHLH, as well as cooperation between the central Orange domain and the C-terminal WRPW motif, is required for transcriptional repressor activity.
Research Fields
· Environmental Information Processing > Signal transduction > Notch signaling pathway. (View pathway)
· Genetic Information Processing > Replication and repair > Fanconi anemia pathway.
· Human Diseases > Endocrine and metabolic diseases > Maturity onset diabetes of the young.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Breast cancer. (View pathway)
References
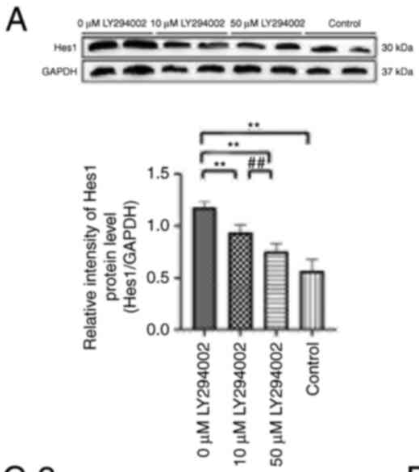
Application: WB Species: Human Sample: SUNE1 cells
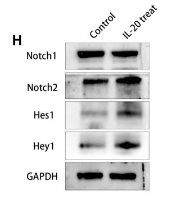
Application: WB Species: Human Sample:
Application: WB Species: mouse Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.