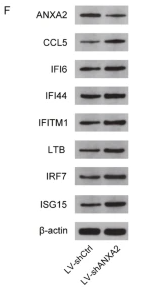
IRF7 Antibody - #DF7503
| Product: | IRF7 Antibody |
| Catalog: | DF7503 |
| Description: | Rabbit polyclonal antibody to IRF7 |
| Application: | WB IHC |
| Cited expt.: | WB, IHC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Horse |
| Mol.Wt.: | 50~70kD; 54kD(Calculated). |
| Uniprot: | Q92985 |
| RRID: | AB_2841003 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF7503, RRID:AB_2841003.
Fold/Unfold
IMD39; Interferon regulatory factor 7; Interferon regulatory factor 7H; IRF 7; IRF 7A; IRF 7H; IRF-7; IRF7; IRF7_HUMAN; IRF7A; IRF7B; IRF7C; IRF7H;
Immunogens
A synthesized peptide derived from human IRF7, corresponding to a region within C-terminal amino acids.
- Q92985 IRF7_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MALAPERAAPRVLFGEWLLGEISSGCYEGLQWLDEARTCFRVPWKHFARKDLSEADARIFKAWAVARGRWPPSSRGGGPPPEAETAERAGWKTNFRCALRSTRRFVMLRDNSGDPADPHKVYALSRELCWREGPGTDQTEAEAPAAVPPPQGGPPGPFLAHTHAGLQAPGPLPAPAGDKGDLLLQAVQQSCLADHLLTASWGADPVPTKAPGEGQEGLPLTGACAGGPGLPAGELYGWAVETTPSPGPQPAALTTGEAAAPESPHQAEPYLSPSPSACTAVQEPSPGALDVTIMYKGRTVLQKVVGHPSCTFLYGPPDPAVRATDPQQVAFPSPAELPDQKQLRYTEELLRHVAPGLHLELRGPQLWARRMGKCKVYWEVGGPPGSASPSTPACLLPRNCDTPIFDFRVFFQELVEFRARQRRGSPRYTIYLGFGQDLSAGRPKEKSLVLVKLEPWLCRVHLEGTQREGVSSLDSSSLSLCLSSANSLYDDIECFLMELEQPA
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Key transcriptional regulator of type I interferon (IFN)-dependent immune responses and plays a critical role in the innate immune response against DNA and RNA viruses. Regulates the transcription of type I IFN genes (IFN-alpha and IFN-beta) and IFN-stimulated genes (ISG) by binding to an interferon-stimulated response element (ISRE) in their promoters. Can efficiently activate both the IFN-beta (IFNB) and the IFN-alpha (IFNA) genes and mediate their induction via both the virus-activated, MyD88-independent pathway and the TLR-activated, MyD88-dependent pathway. Induces transcription of ubiquitin hydrolase USP25 mRNA in response to lipopolysaccharide (LPS) or viral infection in a type I IFN-dependent manner (By similarity). Required during both the early and late phases of the IFN gene induction but is more critical for the late than for the early phase. Exists in an inactive form in the cytoplasm of uninfected cells and following viral infection, double-stranded RNA (dsRNA), or toll-like receptor (TLR) signaling, becomes phosphorylated by IKBKE and TBK1 kinases. This induces a conformational change, leading to its dimerization and nuclear localization where along with other coactivators it can activate transcription of the type I IFN and ISG genes. Can also play a role in regulating adaptive immune responses by inducing PSMB9/LMP2 expression, either directly or through induction of IRF1. Binds to the Q promoter (Qp) of EBV nuclear antigen 1 a (EBNA1) and may play a role in the regulation of EBV latency. Can activate distinct gene expression programs in macrophages and regulate the anti-tumor properties of primary macrophages.
Acetylation inhibits its DNA-binding ability and activity.
In response to a viral infection, phosphorylated on Ser-477 and Ser-479 by TBK1 and IKBKE1. Phosphorylation, and subsequent activation is inhibited by vaccinia virus protein E3. In TLR7- and TLR9-mediated signaling pathway, phosphorylated by IRAK1.
TRAF6-mediated ubiquitination is required for IRF7 activation.
Sumoylated by TRIM28, which inhibits its transactivation activity.
(Microbial infection) Cleaved and inactivated by the protease 3C of enterovirus 71 allowing the virus to disrupt the host type I interferon production.
(Microbial infection) Cleaved and inactivated by the protease 3C of human enterovirus 68D (EV68) allowing the virus to disrupt the host type I interferon production.
Nucleus. Cytoplasm.
Note: The phosphorylated and active form accumulates selectively in the nucleus.
Expressed predominantly in spleen, thymus and peripheral blood leukocytes.
Belongs to the IRF family.
Research Fields
· Human Diseases > Infectious diseases: Viral > Hepatitis C.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Measles.
· Human Diseases > Infectious diseases: Viral > Influenza A.
· Human Diseases > Infectious diseases: Viral > Herpes simplex infection.
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Organismal Systems > Immune system > Toll-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > RIG-I-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Cytosolic DNA-sensing pathway. (View pathway)
References
Application: IHC Species: Mouse Sample: lung tissue
Application: WB Species: Mouse Sample:
Application: WB Species: mouse Sample: lung
Application: WB Species: Human Sample: HK2 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.