FGFR3 Antibody - #AF0160
| Product: | FGFR3 Antibody |
| Catalog: | AF0160 |
| Description: | Rabbit polyclonal antibody to FGFR3 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Zebrafish, Bovine, Sheep, Dog, Chicken |
| Mol.Wt.: | 95kDa; 88kD(Calculated). |
| Uniprot: | P22607 |
| RRID: | AB_2833341 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0160, RRID:AB_2833341.
Fold/Unfold
ACH; CD 333; CD333; CD333 antigen; CEK 2; CEK2; FGFR 3; FGFR-3; FGFR3; FGFR3_HUMAN; Fibroblast growth factor receptor 3 (achondroplasia thanatophoric dwarfism); Fibroblast growth factor receptor 3; Heparin binding growth factor receptor; HSFGFR3EX; Hydroxyaryl protein kinase; JTK 4; JTK4; MFR 3; SAM 3; Tyrosine kinase JTK 4; Tyrosine kinase JTK4; Z FGFR 3;
Immunogens
A synthesized peptide derived from human FGFR3, corresponding to a region within C-terminal amino acids.
Expressed in brain, kidney and testis. Very low or no expression in spleen, heart, and muscle. In 20- to 22-week old fetuses it is expressed at high level in kidney, lung, small intestine and brain, and to a lower degree in spleen, liver, and muscle. Isoform 2 is detected in epithelial cells. Isoform 1 is not detected in epithelial cells. Isoform 1 and isoform 2 are detected in fibroblastic cells.
- P22607 FGFR3_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGAPACALALCVAVAIVAGASSESLGTEQRVVGRAAEVPGPEPGQQEQLVFGSGDAVELSCPPPGGGPMGPTVWVKDGTGLVPSERVLVGPQRLQVLNASHEDSGAYSCRQRLTQRVLCHFSVRVTDAPSSGDDEDGEDEAEDTGVDTGAPYWTRPERMDKKLLAVPAANTVRFRCPAAGNPTPSISWLKNGREFRGEHRIGGIKLRHQQWSLVMESVVPSDRGNYTCVVENKFGSIRQTYTLDVLERSPHRPILQAGLPANQTAVLGSDVEFHCKVYSDAQPHIQWLKHVEVNGSKVGPDGTPYVTVLKTAGANTTDKELEVLSLHNVTFEDAGEYTCLAGNSIGFSHHSAWLVVLPAEEELVEADEAGSVYAGILSYGVGFFLFILVVAAVTLCRLRSPPKKGLGSPTVHKISRFPLKRQVSLESNASMSSNTPLVRIARLSSGEGPTLANVSELELPADPKWELSRARLTLGKPLGEGCFGQVVMAEAIGIDKDRAAKPVTVAVKMLKDDATDKDLSDLVSEMEMMKMIGKHKNIINLLGACTQGGPLYVLVEYAAKGNLREFLRARRPPGLDYSFDTCKPPEEQLTFKDLVSCAYQVARGMEYLASQKCIHRDLAARNVLVTEDNVMKIADFGLARDVHNLDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLLWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTHDLYMIMRECWHAAPSQRPTFKQLVEDLDRVLTVTSTDEYLDLSAPFEQYSPGGQDTPSSSSSGDDSVFAHDLLPPAPPSSGGSRT
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Tyrosine-protein kinase that acts as cell-surface receptor for fibroblast growth factors and plays an essential role in the regulation of cell proliferation, differentiation and apoptosis. Plays an essential role in the regulation of chondrocyte differentiation, proliferation and apoptosis, and is required for normal skeleton development. Regulates both osteogenesis and postnatal bone mineralization by osteoblasts. Promotes apoptosis in chondrocytes, but can also promote cancer cell proliferation. Required for normal development of the inner ear. Phosphorylates PLCG1, CBL and FRS2. Ligand binding leads to the activation of several signaling cascades. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate. Phosphorylation of FRS2 triggers recruitment of GRB2, GAB1, PIK3R1 and SOS1, and mediates activation of RAS, MAPK1/ERK2, MAPK3/ERK1 and the MAP kinase signaling pathway, as well as of the AKT1 signaling pathway. Plays a role in the regulation of vitamin D metabolism. Mutations that lead to constitutive kinase activation or impair normal FGFR3 maturation, internalization and degradation lead to aberrant signaling. Over-expressed or constitutively activated FGFR3 promotes activation of PTPN11/SHP2, STAT1, STAT5A and STAT5B. Secreted isoform 3 retains its capacity to bind FGF1 and FGF2 and hence may interfere with FGF signaling.
Autophosphorylated. Binding of FGF family members together with heparan sulfate proteoglycan or heparin promotes receptor dimerization and autophosphorylation on tyrosine residues. Autophosphorylation occurs in trans between the two FGFR molecules present in the dimer. Phosphorylation at Tyr-724 is essential for stimulation of cell proliferation and activation of PIK3R1, STAT1 and MAP kinase signaling. Phosphorylation at Tyr-760 is required for interaction with PIK3R1 and PLCG1.
Ubiquitinated. Is rapidly ubiquitinated after ligand binding and autophosphorylation, leading to receptor internalization and degradation. Subject to both proteasomal and lysosomal degradation.
N-glycosylated in the endoplasmic reticulum. The N-glycan chains undergo further maturation to an Endo H-resistant form in the Golgi apparatus.
Cell membrane>Single-pass type I membrane protein. Cytoplasmic vesicle. Endoplasmic reticulum.
Note: The activated receptor is rapidly internalized and degraded. Detected in intracellular vesicles after internalization of the autophosphorylated receptor.
Cell membrane>Single-pass type I membrane protein.
Secreted.
Cell membrane>Single-pass type I membrane protein.
Expressed in brain, kidney and testis. Very low or no expression in spleen, heart, and muscle. In 20- to 22-week old fetuses it is expressed at high level in kidney, lung, small intestine and brain, and to a lower degree in spleen, liver, and muscle. Isoform 2 is detected in epithelial cells. Isoform 1 is not detected in epithelial cells. Isoform 1 and isoform 2 are detected in fibroblastic cells.
The second and third Ig-like domains directly interact with fibroblast growth factors (FGF) and heparan sulfate proteoglycans.
Belongs to the protein kinase superfamily. Tyr protein kinase family. Fibroblast growth factor receptor subfamily.
Research Fields
· Cellular Processes > Transport and catabolism > Endocytosis. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Signaling pathways regulating pluripotency of stem cells. (View pathway)
· Cellular Processes > Cell motility > Regulation of actin cytoskeleton. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Ras signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Rap1 signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > EGFR tyrosine kinase inhibitor resistance.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Bladder cancer. (View pathway)
· Human Diseases > Cancers: Overview > Central carbon metabolism in cancer. (View pathway)
References
Application: IF/ICC Species: Mouse Sample:
Application: WB Species: mice Sample: RAW264.7 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.








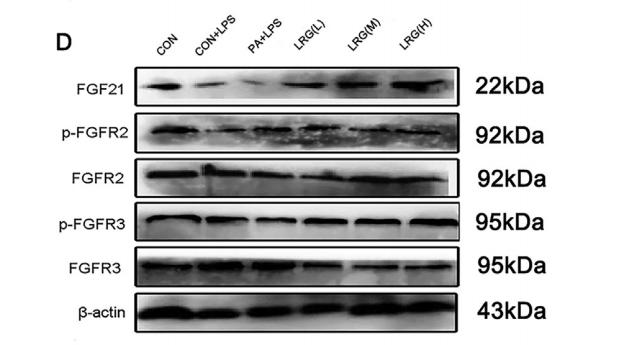
![Figure 3 FOXG1 binds and regulates the expression of Fgfr3. (A) RNA-seq analysis of FACS-purified Control and Foxg1 loss-of-function (LOF) progenitors harvested two days after labelling at E15.5. Gliogenic factors such as Nfia, Id3, and Olig3 are upregulated, and neuronal markers such as Pou3f1 and Robo4 are downregulated. (B–D) Multimodal analysis comparing FOXG1 occupancy (ChIP-seq) and bivalent epigenetic marks (H3K4Me3 and H3K27Me3) and astrocyte-enriched genes from Telley et al., 2019 reveals a list of 19 genes common to each dataset (B). Four of these are upregulated upon loss of Foxg1, including the known gliogenic gene Fgfr3 (D). FOXG1 occupies a –26 kb enhancer region of Fgfr3 (C). (E) KEGG analysis of the upregulated genes from (A) identifies the MAPK signalling pathway downstream of FGF signalling. (F) Loss of Foxg1 from E15.5 progenitors (hGFAP-CreERT2, tamoxifen at E15.5) causes upregulation of FGFR3 receptor by E17.5 as seen in cells near the VZ of the somatosensory cortex. Boxes (F) indicate the regions in high magnification shown in the adjacent panels (G). Dashed circles outline the regions of interest (ROIs) identified in the DAPI channel used for intensity quantification in (H). (G; n = 50 [Control and Foxg1 LOF] ROIs from N = 3 brains) and phosphorylated-ERK1/2 (H; n = 67 [Control] and 89 [Foxg1 LOF]) cells from N = 3 brains (biologically independent replicates). Statistical test: Mann–Whitney test *p FGFR3 Antibody - Figure 3 FOXG1 binds and regulates the expression of Fgfr3.](http://img.affbiotech.cn/uploads/202509/a653b865d39e0c063e2bc9ede5febfd9.png)