PI 3 Kinase Class 2A Antibody - #DF2623
| Product: | PI 3 Kinase Class 2A Antibody |
| Catalog: | DF2623 |
| Description: | Rabbit polyclonal antibody to PI 3 Kinase Class 2A |
| Application: | WB |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse |
| Prediction: | Pig, Zebrafish, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 191 kDa; 191kD(Calculated). |
| Uniprot: | O00443 |
| RRID: | AB_2839829 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF2623, RRID:AB_2839829.
Fold/Unfold
C2 containing phosphatidylinositol kinase; C2 domain containing alpha polypeptide; Class 2 alpha polypeptide; CPK; Cpk m; DKFZp686L193; MGC142218; OTTHUMP00000231763; P3C2A_HUMAN; Phosphatidylinositol 3 kinase class 2 alpha; Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing subunit alpha; Phosphoinositide 3 kinase class 2 alpha; Phosphoinositide 3 kinase, class 2, alpha polypeptide; Phosphoinositide 3-Kinase-C2-alpha; PI3-K-C2(ALPHA); PI3-K-C2A; PI3K-C2-alpha; PI3K-C2alpha; PI3KC 2 alpha; PIK3C 2A; Pik3c2a; PtdIns-3-kinase C2 subunit alpha;
Immunogens
A synthesized peptide derived from human PI 3 Kinase Class 2A, corresponding to a region within C-terminal amino acids.
Expressed in columnar and transitional epithelia, mononuclear cells, smooth muscle cells, and endothelial cells lining capillaries and small venules (at protein level). Ubiquitously expressed, with highest levels in heart, placenta and ovary, and lowest levels in the kidney. Detected at low levels in islets of Langerhans from type 2 diabetes mellitus individuals.
- O00443 P3C2A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAQISSNSGFKECPSSHPEPTRAKDVDKEEALQMEAEALAKLQKDRQVTDNQRGFELSSSTRKKAQVYNKQDYDLMVFPESDSQKRALDIDVEKLTQAELEKLLLDDSFETKKTPVLPVTPILSPSFSAQLYFRPTIQRGQWPPGLPGPSTYALPSIYPSTYSKQAAFQNGFNPRMPTFPSTEPIYLSLPGQSPYFSYPLTPATPFHPQGSLPIYRPVVSTDMAKLFDKIASTSEFLKNGKARTDLEITDSKVSNLQVSPKSEDISKFDWLDLDPLSKPKVDNVEVLDHEEEKNVSSLLAKDPWDAVLLEERSTANCHLERKVNGKSLSVATVTRSQSLNIRTTQLAKAQGHISQKDPNGTSSLPTGSSLLQEVEVQNEEMAAFCRSITKLKTKFPYTNHRTNPGYLLSPVTAQRNICGENASVKVSIDIEGFQLPVTFTCDVSSTVEIIIMQALCWVHDDLNQVDVGSYVLKVCGQEEVLQNNHCLGSHEHIQNCRKWDTEIRLQLLTFSAMCQNLARTAEDDETPVDLNKHLYQIEKPCKEAMTRHPVEELLDSYHNQVELALQIENQHRAVDQVIKAVRKICSALDGVETLAITESVKKLKRAVNLPRSKTADVTSLFGGEDTSRSSTRGSLNPENPVQVSINQLTAAIYDLLRLHANSGRSPTDCAQSSKSVKEAWTTTEQLQFTIFAAHGISSNWVSNYEKYYLICSLSHNGKDLFKPIQSKKVGTYKNFFYLIKWDELIIFPIQISQLPLESVLHLTLFGILNQSSGSSPDSNKQRKGPEALGKVSLPLFDFKRFLTCGTKLLYLWTSSHTNSVPGTVTKKGYVMERIVLQVDFPSPAFDIIYTTPQVDRSIIQQHNLETLENDIKGKLLDILHKDSSLGLSKEDKAFLWEKRYYCFKHPNCLPKILASAPNWKWVNLAKTYSLLHQWPALYPLIALELLDSKFADQEVRSLAVTWIEAISDDELTDLLPQFVQALKYEIYLNSSLVQFLLSRALGNIQIAHNLYWLLKDALHDVQFSTRYEHVLGALLSVGGKRLREELLKQTKLVQLLGGVAEKVRQASGSARQVVLQRSMERVQSFFQKNKCRLPLKPSLVAKELNIKSCSFFSSNAVPLKVTMVNADPMGEEINVMFKVGEDLRQDMLALQMIKIMDKIWLKEGLDLRMVIFKCLSTGRDRGMVELVPASDTLRKIQVEYGVTGSFKDKPLAEWLRKYNPSEEEYEKASENFIYSCAGCCVATYVLGICDRHNDNIMLRSTGHMFHIDFGKFLGHAQMFGSFKRDRAPFVLTSDMAYVINGGEKPTIRFQLFVDLCCQAYNLIRKQTNLFLNLLSLMIPSGLPELTSIQDLKYVRDALQPQTTDAEATIFFTRLIESSLGSIATKFNFFIHNLAQLRFSGLPSNDEPILSFSPKTYSFRQDGRIKEVSVFTYHKKYNPDKHYIYVVRILREGQIEPSFVFRTFDEFQELHNKLSIIFPLWKLPGFPNRMVLGRTHIKDVAAKRKIELNSYLQSLMNASTDVAECDLVCTFFHPLLRDEKAEGIARSADAGSFSPTPGQIGGAVKLSISYRNGTLFIMVMHIKDLVTEDGADPNPYVKTYLLPDNHKTSKRKTKISRKTRNPTFNEMLVYSGYSKETLRQRELQLSVLSAESLRENFFLGGVTLPLKDFNLSKETVKWYQLTAATYL
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Generates phosphatidylinositol 3-phosphate (PtdIns3P) and phosphatidylinositol 3,4-bisphosphate (PtdIns(3,4)P2) that act as second messengers. Has a role in several intracellular trafficking events. Functions in insulin signaling and secretion. Required for translocation of the glucose transporter SLC2A4/GLUT4 to the plasma membrane and glucose uptake in response to insulin-mediated RHOQ activation. Regulates insulin secretion through two different mechanisms: involved in glucose-induced insulin secretion downstream of insulin receptor in a pathway that involves AKT1 activation and TBC1D4/AS160 phosphorylation, and participates in the late step of insulin granule exocytosis probably in insulin granule fusion. Synthesizes PtdIns3P in response to insulin signaling. Functions in clathrin-coated endocytic vesicle formation and distribution. Regulates dynamin-independent endocytosis, probably by recruiting EEA1 to internalizing vesicles. In neurosecretory cells synthesizes PtdIns3P on large dense core vesicles. Participates in calcium induced contraction of vascular smooth muscle by regulating myosin light chain (MLC) phosphorylation through a mechanism involving Rho kinase-dependent phosphorylation of the MLCP-regulatory subunit MYPT1. May play a role in the EGF signaling cascade. May be involved in mitosis and UV-induced damage response. Required for maintenance of normal renal structure and function by supporting normal podocyte function. Involved in the regulation of ciliogenesis and trafficking of ciliary components.
Phosphorylated upon insulin stimulation; which may lead to enzyme activation (By similarity). Phosphorylated on Ser-259 during mitosis and upon UV irradiation; which does not change enzymatic activity but leads to proteasomal degradation. Ser-259 phosphorylation may be mediated by CDK1 or JNK, depending on the physiological state of the cell.
Cell membrane. Golgi apparatus. Cytoplasmic vesicle>Clathrin-coated vesicle. Nucleus. Cytoplasm.
Note: Inserts preferentially into membranes containing PtdIns(4,5)P2 (PubMed:17038310). Associated with RNA-containing structures (PubMed:11606566).
Expressed in columnar and transitional epithelia, mononuclear cells, smooth muscle cells, and endothelial cells lining capillaries and small venules (at protein level). Ubiquitously expressed, with highest levels in heart, placenta and ovary, and lowest levels in the kidney. Detected at low levels in islets of Langerhans from type 2 diabetes mellitus individuals.
Belongs to the PI3/PI4-kinase family.
Research Fields
· Environmental Information Processing > Signal transduction > Phosphatidylinositol signaling system.
· Metabolism > Carbohydrate metabolism > Inositol phosphate metabolism.
· Metabolism > Global and overview maps > Metabolic pathways.
References
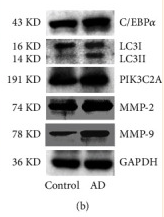
Application: WB Species: Rat Sample: aorta tissues
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.