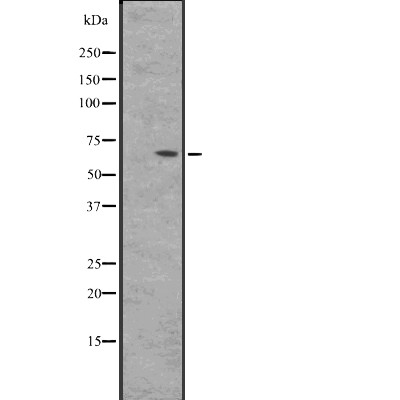
BIN2 Antibody - #DF2479
| Product: | BIN2 Antibody |
| Catalog: | DF2479 |
| Description: | Rabbit polyclonal antibody to BIN2 |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human |
| Mol.Wt.: | 62 kDa; 62kD(Calculated). |
| Uniprot: | Q9UBW5 |
| RRID: | AB_2839685 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF2479, RRID:AB_2839685.
Fold/Unfold
BIN2; BIN2_HUMAN; BRAP 1; Breast cancer associated protein BRAP1; Breast cancer-associated protein 1; Bridging integrator 2;
Immunogens
A synthesized peptide derived from human BIN2, corresponding to a region within the internal amino acids.
Detected in natural killer cells (at protein level). Highest level expression seen in spleen and peripheral blood leukocytes and is also expressed at high levels in thymus, colon and placenta, suggesting preferential expression in hematopoietic tissues.
- Q9UBW5 BIN2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAEGKAGGAAGLFAKQVQKKFSRAQEKVLQKLGKAVETKDERFEQSASNFYQQQAEGHKLYKDLKNFLSAVKVMHESSKRVSETLQEIYSSEWDGHEELKAIVWNNDLLWEDYEEKLADQAVRTMEIYVAQFSEIKERIAKRGRKLVDYDSARHHLEAVQNAKKKDEAKTAKAEEEFNKAQTVFEDLNQELLEELPILYNSRIGCYVTIFQNISNLRDVFYREMSKLNHNLYEVMSKLEKQHSNKVFVVKGLSSSSRRSLVISPPVRTATVSSPLTSPTSPSTLSLKSESESVSATEDLAPDAAQGEDNSEIKELLEEEEIEKEGSEASSSEEDEPLPACNGPAQAQPSPTTERAKSQEEVLPSSTTPSPGGALSPSGQPSSSATEVVLRTRTASEGSEQPKKRASIQRTSAPPSRPPPPRATASPRPSSGNIPSSPTASGGGSPTSPRASLGTGTASPRTSLEVSPNPEPPEKPVRTPEAKENENIHNQNPEELCTSPTLMTSQVASEPGEAKKMEDKEKDNKLISANSSEGQDQLQVSMVPENNNLTAPEPQEEVSTSENPQL
Research Backgrounds
Promotes cell motility and migration, probably via its interaction with the cell membrane and with podosome proteins that mediate interaction with the cytoskeleton. Modulates membrane curvature and mediates membrane tubulation. Plays a role in podosome formation. Inhibits phagocytosis.
Cytoplasm. Cell projection>Podosome membrane>Peripheral membrane protein>Cytoplasmic side. Cytoplasm>Cell cortex. Cell projection>Phagocytic cup.
Note: Associates with membranes enriched in phosphoinositides. Detected in the actin-rich cell cortex at the leading edge of migrating cells. Detected at podosomes, at an actin-rich ring-like structure.
Detected in natural killer cells (at protein level). Highest level expression seen in spleen and peripheral blood leukocytes and is also expressed at high levels in thymus, colon and placenta, suggesting preferential expression in hematopoietic tissues.
The BAR domain mediates dimerization and interaction with membranes enriched in phosphatidylinositides.
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.