CD2AP Antibody - #DF2298
| Product: | CD2AP Antibody |
| Catalog: | DF2298 |
| Description: | Rabbit polyclonal antibody to CD2AP |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken |
| Mol.Wt.: | 74 kDa; 71kD(Calculated). |
| Uniprot: | Q9Y5K6 |
| RRID: | AB_2839524 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF2298, RRID:AB_2839524.
Fold/Unfold
Adapter protein CMS; AL024079; C78928; Cas ligand with multiple SH3 domains; CD2 associated protein; CD2-associated protein; CD2AP; CD2AP_HUMAN; CMS; Mesenchyme to epithelium transition protein with SH3 domains 1; METS 1; Mets1;
Immunogens
A synthesized peptide derived from human CD2AP, corresponding to a region within the internal amino acids.
- Q9Y5K6 CD2AP_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MVDYIVEYDYDAVHDDELTIRVGEIIRNVKKLQEEGWLEGELNGRRGMFPDNFVKEIKRETEFKDDSLPIKRERHGNVASLVQRISTYGLPAGGIQPHPQTKNIKKKTKKRQCKVLFEYIPQNEDELELKVGDIIDINEEVEEGWWSGTLNNKLGLFPSNFVKELEVTDDGETHEAQDDSETVLAGPTSPIPSLGNVSETASGSVTQPKKIRGIGFGDIFKEGSVKLRTRTSSSETEEKKPEKPLILQSLGPKTQSVEITKTDTEGKIKAKEYCRTLFAYEGTNEDELTFKEGEIIHLISKETGEAGWWRGELNGKEGVFPDNFAVQINELDKDFPKPKKPPPPAKAPAPKPELIAAEKKYFSLKPEEKDEKSTLEQKPSKPAAPQVPPKKPTPPTKASNLLRSSGTVYPKRPEKPVPPPPPIAKINGEVSSISSKFETEPVSKLKLDSEQLPLRPKSVDFDSLTVRTSKETDVVNFDDIASSENLLHLTANRPKMPGRRLPGRFNGGHSPTHSPEKILKLPKEEDSANLKPSELKKDTCYSPKPSVYLSTPSSASKANTTAFLTPLEIKAKVETDDVKKNSLDELRAQIIELLCIVEALKKDHGKELEKLRKDLEEEKTMRSNLEMEIEKLKKAVLSS
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Seems to act as an adapter protein between membrane proteins and the actin cytoskeleton. In collaboration with CBLC, modulates the rate of RET turnover and may act as regulatory checkpoint that limits the potency of GDNF on neuronal survival. Controls CBLC function, converting it from an inhibitor to a promoter of RET degradation (By similarity). May play a role in receptor clustering and cytoskeletal polarity in the junction between T-cell and antigen-presenting cell (By similarity). May anchor the podocyte slit diaphragm to the actin cytoskeleton in renal glomerolus. Also required for cytokinesis. Plays a role in epithelial cell junctions formation.
Phosphorylated on tyrosine residues; probably by c-Abl, Fyn and c-Src.
Cytoplasm>Cytoskeleton. Cell projection>Ruffle. Cell junction.
Note: Colocalizes with F-actin and BCAR1/p130Cas in membrane ruffles (PubMed:10339567). Located at podocyte slit diaphragm between podocyte foot processes (By similarity). During late anaphase and telophase, concentrates in the vicinity of the midzone microtubules and in the midbody in late telophase (PubMed:15800069).
Widely expressed in fetal and adult tissues.
The Pro-rich domain may mediate binding to SH3 domains.
Potential homodimerization is mediated by the coiled coil domain.
Research Fields
· Human Diseases > Infectious diseases: Bacterial > Bacterial invasion of epithelial cells.
References
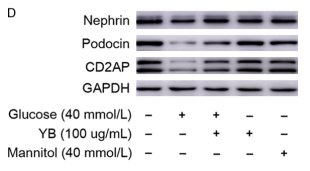
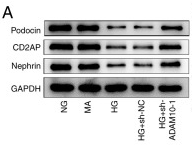
Application: WB Species: Mouse Sample:
Application: WB Species: Human Sample: podocytes
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.