Tubulin beta Antibody - #AF7011
| Product: | Tubulin beta Antibody |
| Catalog: | AF7011 |
| Description: | Rabbit polyclonal antibody to Tubulin beta |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Mol.Wt.: | 55kDa; 50kD(Calculated). |
| Uniprot: | P07437 |
| RRID: | AB_2827688 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF7011, RRID:AB_2827688.
Fold/Unfold
TUBB3, CDCBM, Beta III Tubulin, Class III beta-tubulin, TUBB4, Tubulin, beta 3, Tubulin beta-III, Tubulin beta-3 chain, Tubulin beta-4 chain, Tubulin, beta 3 class III, CFEOM3A
Immunogens
A synthesized peptide derived from human Tubulin beta.
- P07437 TBB5_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MREIVHIQAGQCGNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLDRISVYYNEATGGKYVPRAILVDLEPGTMDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVVRKEAESCDCLQGFQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQLVENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQVFDAKNMMAACDPRHGRYLTVAAVFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMAVTFIGNSTAIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATAEEEEDFGEEAEEEA
Research Backgrounds
Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
Some glutamate residues at the C-terminus are polyglutamylated, resulting in polyglutamate chains on the gamma-carboxyl group. Polyglutamylation plays a key role in microtubule severing by spastin (SPAST). SPAST preferentially recognizes and acts on microtubules decorated with short polyglutamate tails: severing activity by SPAST increases as the number of glutamates per tubulin rises from one to eight, but decreases beyond this glutamylation threshold.
Some glutamate residues at the C-terminus are monoglycylated but not polyglycylated due to the absence of functional TTLL10 in human. Monoglycylation is mainly limited to tubulin incorporated into axonemes (cilia and flagella). Both polyglutamylation and monoglycylation can coexist on the same protein on adjacent residues, and lowering glycylation levels increases polyglutamylation, and reciprocally. The precise function of monoglycylation is still unclear (Probable).
Phosphorylated on Ser-172 by CDK1 during the cell cycle, from metaphase to telophase, but not in interphase. This phosphorylation inhibits tubulin incorporation into microtubules.
Cytoplasm>Cytoskeleton.
Ubiquitously expressed with highest levels in spleen, thymus and immature brain.
The highly acidic C-terminal region may bind cations such as calcium.
Belongs to the tubulin family.
Research Fields
· Cellular Processes > Transport and catabolism > Phagosome. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Gap junction. (View pathway)
· Human Diseases > Infectious diseases: Bacterial > Pathogenic Escherichia coli infection.
References
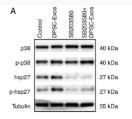
Application: WB Species: human Sample: HEK293 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.






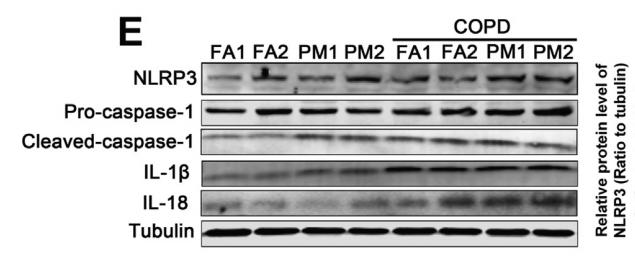
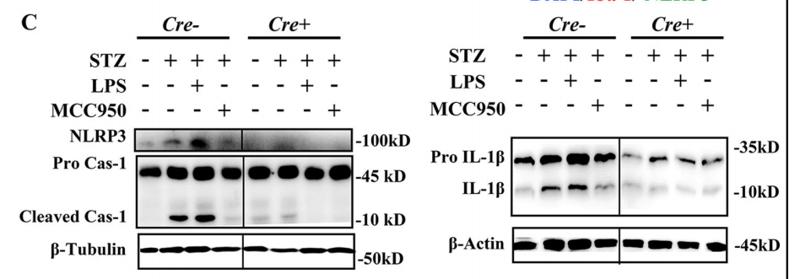










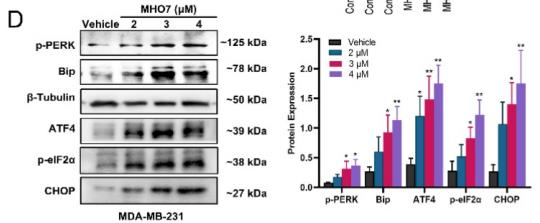
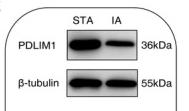











![Fig. 1. XAF1 facilitates antiviral immune signaling. (A) Flow chart depicting the process of MAVS-TurboID proximity-based labeling technology to detect interacting proteins. XAF1 was identified as a MAVS-interacting protein by mass spectrometry. (B) Luciferase (Luci) activity in HEK293T cells transiently transfected with 50 ng of the pRL-TK reporter, 50 ng of luciferase reporter driven by promoters of genes encoding IFN-β, empty vector (Vector), or XAF1 together with expression plasmids for Cgas, STING, cGAS+STING, N-MDA5, N-RIG-I, MAVS, or TBK1 for 24 hours. (C and D) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of IFNB1 and ISG15 was performed after NT (nontreated) or after stimulation with SeV, VSV, or HSV-1 for 12 hours in XAF1−/− HT-29 (C) and THP-1 (D) cells. (E and F) 2fTGH-ISRE (IFN-stimulated response element) cells were treated with supernatant from control and XAF1−/− HT-29 (E) and THP-1 (F) cells, which were NT or stimulated with SeV, VSV, or HSV-1 for 12 hours. After 6 hours of treatment of the supernatant, ISRE luciferase activity was measured. (G) Heatmap for selected ISGs in WT and XAF1−/− HT-29 cells treated with or without VSV for 6 hours. (H) Histogram of the combined score (https://david.ncifcrf.gov/tools.jsp) for biological processes for the up-regulated genes in WT versus XAF1−/− HT-29 cells treated with or without VSV for 6 hours. Data are the means ± SEM (n = 3) from three independent experiments; NS, not significant; *P < 0.05 and **P < 0.01 [Student’s t test, (B) to (F)]. Data are representative of two [(G) and (H)] or three [(B) to (F)] independent experiments with similar results. Tubulin beta Antibody - Fig.](http://img.affbiotech.cn/uploads/202412/f2c121f800e8ec18fac417d81a307b02.png)




