DNMT3A Antibody - #DF7226
| Product: | DNMT3A Antibody |
| Catalog: | DF7226 |
| Description: | Rabbit polyclonal antibody to DNMT3A |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Rabbit, Dog |
| Mol.Wt.: | 120-140kD; 102kD(Calculated). |
| Uniprot: | Q9Y6K1 |
| RRID: | AB_2839167 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF7226, RRID:AB_2839167.
Fold/Unfold
DNA (cytosine 5) methyltransferase 3 alpha; DNA (cytosine 5) methyltransferase 3A; DNA (cytosine-5)-methyltransferase 3A; DNA cytosine methyltransferase 3A2; DNA methyltransferase 3 alpha; DNA methyltransferase 3a; DNA methyltransferase HsaIIIA; DNA MTase HsaIIIA; DNM3A_HUMAN; DNMT 3a; DNMT; Dnmt3a; DNMT3A2; M.HsaIIIA; MCMT; OTTHUMP00000201149; TBRS;
Immunogens
A synthesized peptide derived from human DNMT3A, corresponding to a region within the internal amino acids.
Highly expressed in fetal tissues, skeletal muscle, heart, peripheral blood mononuclear cells, kidney, and at lower levels in placenta, brain, liver, colon, spleen, small intestine and lung.
- Q9Y6K1 DNM3A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MPAMPSSGPGDTSSSAAEREEDRKDGEEQEEPRGKEERQEPSTTARKVGRPGRKRKHPPVESGDTPKDPAVISKSPSMAQDSGASELLPNGDLEKRSEPQPEEGSPAGGQKGGAPAEGEGAAETLPEASRAVENGCCTPKEGRGAPAEAGKEQKETNIESMKMEGSRGRLRGGLGWESSLRQRPMPRLTFQAGDPYYISKRKRDEWLARWKREAEKKAKVIAGMNAVEENQGPGESQKVEEASPPAVQQPTDPASPTVATTPEPVGSDAGDKNATKAGDDEPEYEDGRGFGIGELVWGKLRGFSWWPGRIVSWWMTGRSRAAEGTRWVMWFGDGKFSVVCVEKLMPLSSFCSAFHQATYNKQPMYRKAIYEVLQVASSRAGKLFPVCHDSDESDTAKAVEVQNKPMIEWALGGFQPSGPKGLEPPEEEKNPYKEVYTDMWVEPEAAAYAPPPPAKKPRKSTAEKPKVKEIIDERTRERLVYEVRQKCRNIEDICISCGSLNVTLEHPLFVGGMCQNCKNCFLECAYQYDDDGYQSYCTICCGGREVLMCGNNNCCRCFCVECVDLLVGPGAAQAAIKEDPWNCYMCGHKGTYGLLRRREDWPSRLQMFFANNHDQEFDPPKVYPPVPAEKRKPIRVLSLFDGIATGLLVLKDLGIQVDRYIASEVCEDSITVGMVRHQGKIMYVGDVRSVTQKHIQEWGPFDLVIGGSPCNDLSIVNPARKGLYEGTGRLFFEFYRLLHDARPKEGDDRPFFWLFENVVAMGVSDKRDISRFLESNPVMIDAKEVSAAHRARYFWGNLPGMNRPLASTVNDKLELQECLEHGRIAKFSKVRTITTRSNSIKQGKDQHFPVFMNEKEDILWCTEMERVFGFPVHYTDVSNMSRLARQRLLGRSWSVPVIRHLFAPLKEYFACV
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Required for genome-wide de novo methylation and is essential for the establishment of DNA methylation patterns during development. DNA methylation is coordinated with methylation of histones. It modifies DNA in a non-processive manner and also methylates non-CpG sites. May preferentially methylate DNA linker between 2 nucleosomal cores and is inhibited by histone H1. Plays a role in paternal and maternal imprinting. Required for methylation of most imprinted loci in germ cells. Acts as a transcriptional corepressor for ZBTB18. Recruited to trimethylated 'Lys-36' of histone H3 (H3K36me3) sites. Can actively repress transcription through the recruitment of HDAC activity.
Sumoylated; sumoylation disrupts the ability to interact with histone deacetylases (HDAC1 and HDAC2) and repress transcription.
Nucleus. Cytoplasm.
Note: Accumulates in the major satellite repeats at pericentric heterochromatin.
Highly expressed in fetal tissues, skeletal muscle, heart, peripheral blood mononuclear cells, kidney, and at lower levels in placenta, brain, liver, colon, spleen, small intestine and lung.
The PWWP domain is essential for targeting to pericentric heterochromatin. It specifically recognizes and binds trimethylated 'Lys-36' of histone H3 (H3K36me3) (By similarity).
Belongs to the class I-like SAM-binding methyltransferase superfamily. C5-methyltransferase family.
Research Fields
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Metabolism > Amino acid metabolism > Cysteine and methionine metabolism.
· Metabolism > Global and overview maps > Metabolic pathways.
References
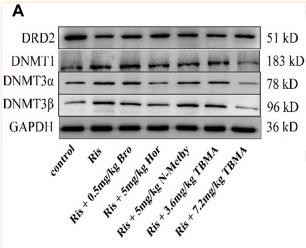
Application: WB Species: Rat Sample:
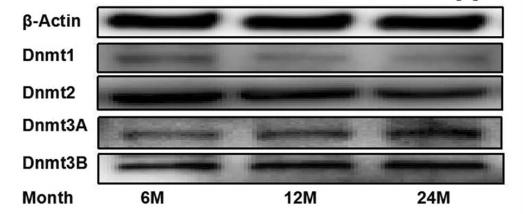
Application: WB Species: Mouse Sample: GCs
Application: WB Species: rat Sample: ovaries
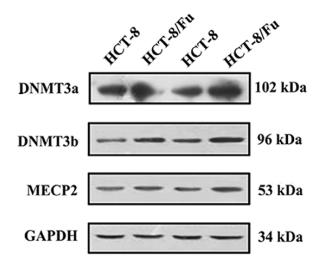
Application: WB Species: human Sample: HCT-8 and HCT-8/Fu cells
Application: WB Species: Rat Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.