Connexin 43 / GJA1 Antibody - #AF0137
| Product: | Connexin 43 / GJA1 Antibody |
| Catalog: | AF0137 |
| Description: | Rabbit polyclonal antibody to Connexin 43 / GJA1 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 43kDa; 43kD(Calculated). |
| Uniprot: | P17302 |
| RRID: | AB_2833319 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0137, RRID:AB_2833319.
Fold/Unfold
Connexin 43; Connexin-43; Cx 43; Cx43; CXA1_HUMAN; DFNB38; Gap junction 43 kDa heart protein; Gap junction alpha-1 protein; Gap junction protein alpha 1 43kDa (connexin 43); Gap junction protein alpha 1 43kDa; Gap junction protein alpha like; GJA 1; Gja1; GJAL; ODD; ODDD; ODOD; SDTY3;
Immunogens
A synthesized peptide derived from human Connexin 43 / GJA1, corresponding to a region within the internal amino acids.
- P17302 CXA1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGDWSALGKLLDKVQAYSTAGGKVWLSVLFIFRILLLGTAVESAWGDEQSAFRCNTQQPGCENVCYDKSFPISHVRFWVLQIIFVSVPTLLYLAHVFYVMRKEEKLNKKEEELKVAQTDGVNVDMHLKQIEIKKFKYGIEEHGKVKMRGGLLRTYIISILFKSIFEVAFLLIQWYIYGFSLSAVYTCKRDPCPHQVDCFLSRPTEKTIFIIFMLVVSLVSLALNIIELFYVFFKGVKDRVKGKSDPYHATSGALSPAKDCGSQKYAYFNGCSSPTAPLSPMSPPGYKLVTGDRNNSSCRNYNKQASEQNWANYSAEQNRMGQAGSTISNSHAQPFDFPDDNQNSKKLAAGHELQPLAIVDQRPSSRASSRASSRPRPDDLEI
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Gap junction protein that acts as a regulator of bladder capacity. A gap junction consists of a cluster of closely packed pairs of transmembrane channels, the connexons, through which materials of low MW diffuse from one cell to a neighboring cell. May play a critical role in the physiology of hearing by participating in the recycling of potassium to the cochlear endolymph. Negative regulator of bladder functional capacity: acts by enhancing intercellular electrical and chemical transmission, thus sensitizing bladder muscles to cholinergic neural stimuli and causing them to contract (By similarity). May play a role in cell growth inhibition through the regulation of NOV expression and localization. Plays an essential role in gap junction communication in the ventricles (By similarity).
Phosphorylated at Ser-368 by PRKCG; phosphorylation induces disassembly of gap junction plaques and inhibition of gap junction activity (By similarity). Phosphorylation at Ser-325, Ser-328 and Ser-330 by CK1 modulates gap junction assembly. Phosphorylation at Ser-368 by PRKCD triggers its internalization into small vesicles leading to proteasome-mediated degradation (By similarity).
Sumoylated with SUMO1, SUMO2 and SUMO3, which may regulate the level of functional Cx43 gap junctions at the plasma membrane. May be desumoylated by SENP1 or SENP2.
S-nitrosylation at Cys-271 is enriched at the muscle endothelial gap junction in arteries, it augments channel permeability and may regulate of smooth muscle cell to endothelial cell communication.
Acetylated in the developing cortex; leading to delocalization from the cell membrane.
Cell membrane>Multi-pass membrane protein. Cell junction>Gap junction. Endoplasmic reticulum.
Note: Localizes at the intercalated disk (ICD) in cardiomyocytes and the proper localization at ICD is dependent on TMEM65.
Expressed in the heart and fetal cochlea.
Belongs to the connexin family. Alpha-type (group II) subfamily.
Research Fields
· Cellular Processes > Cellular community - eukaryotes > Gap junction. (View pathway)
· Human Diseases > Cardiovascular diseases > Arrhythmogenic right ventricular cardiomyopathy (ARVC).
References
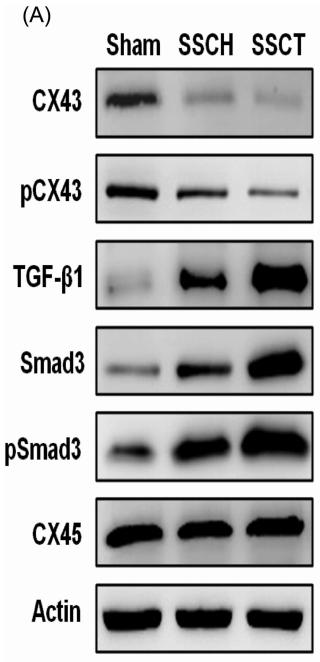
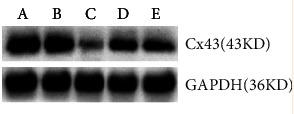
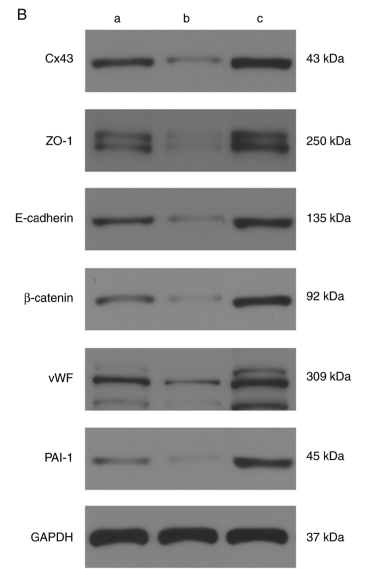
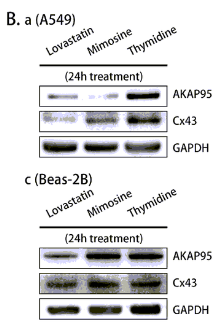
Application: WB Species: Mouse Sample:
Application: IF/ICC Species: Mouse Sample:
Application: IF/ICC Species: rat Sample:
Application: WB Species: Rat Sample:
Application: WB Species: Human Sample: HPMECs
Application: WB Species: rat Sample: sacral spinal cord
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.