ERK5 Antibody - #DF6835
| Product: | ERK5 Antibody |
| Catalog: | DF6835 |
| Description: | Rabbit polyclonal antibody to ERK5 |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Xenopus |
| Mol.Wt.: | 89kDa; 88kD(Calculated). |
| Uniprot: | Q13164 |
| RRID: | AB_2838794 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF6835, RRID:AB_2838794.
Fold/Unfold
Big MAP kinase 1; BMK 1; BMK 1 kinase; BMK-1; BMK1; BMK1 Kinase; EC 2.7.11.24; ERK 4; ERK 5; ERK-5; ERK4; ERK5; Extracellular signal regulated kinase 5; Extracellular signal-regulated kinase 5; MAP kinase 7; MAPK 7; MAPK7; Mitogen activated protein kinase 7; Mitogen-activated protein kinase 7; MK07_HUMAN; OTTHUMP00000065906; OTTHUMP00000065907; PRKM 7; PRKM7; PROTEIN KINASE, MITOGEN-ACTIVATED, 7;
Immunogens
A synthesized peptide derived from human ERK5, corresponding to a region within the internal amino acids.
Expressed in many adult tissues. Abundant in heart, placenta, lung, kidney and skeletal muscle. Not detectable in liver.
- Q13164 MK07_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAEPLKEEDGEDGSAEPPGPVKAEPAHTAASVAAKNLALLKARSFDVTFDVGDEYEIIETIGNGAYGVVSSARRRLTGQQVAIKKIPNAFDVVTNAKRTLRELKILKHFKHDNIIAIKDILRPTVPYGEFKSVYVVLDLMESDLHQIIHSSQPLTLEHVRYFLYQLLRGLKYMHSAQVIHRDLKPSNLLVNENCELKIGDFGMARGLCTSPAEHQYFMTEYVATRWYRAPELMLSLHEYTQAIDLWSVGCIFGEMLARRQLFPGKNYVHQLQLIMMVLGTPSPAVIQAVGAERVRAYIQSLPPRQPVPWETVYPGADRQALSLLGRMLRFEPSARISAAAALRHPFLAKYHDPDDEPDCAPPFDFAFDREALTRERIKEAIVAEIEDFHARREGIRQQIRFQPSLQPVASEPGCPDVEMPSPWAPSGDCAMESPPPAPPPCPGPAPDTIDLTLQPPPPVSEPAPPKKDGAISDNTKAALKAALLKSLRSRLRDGPSAPLEAPEPRKPVTAQERQREREEKRRRRQERAKEREKRRQERERKERGAGASGGPSTDPLAGLVLSDNDRSLLERWTRMARPAAPALTSVPAPAPAPTPTPTPVQPTSPPPGPVAQPTGPQPQSAGSTSGPVPQPACPPPGPAPHPTGPPGPIPVPAPPQIATSTSLLAAQSLVPPPGLPGSSTPGVLPYFPPGLPPPDAGGAPQSSMSESPDVNLVTQQLSKSQVEDPLPPVFSGTPKGSGAGYGVGFDLEEFLNQSFDMGVADGPQDGQADSASLSASLLADWLEGHGMNPADIESLQREIQMDSPMLLADLPDLQDP
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Plays a role in various cellular processes such as proliferation, differentiation and cell survival. The upstream activator of MAPK7 is the MAPK kinase MAP2K5. Upon activation, it translocates to the nucleus and phosphorylates various downstream targets including MEF2C. EGF activates MAPK7 through a Ras-independent and MAP2K5-dependent pathway. May have a role in muscle cell differentiation. May be important for endothelial function and maintenance of blood vessel integrity. MAP2K5 and MAPK7 interact specifically with one another and not with MEK1/ERK1 or MEK2/ERK2 pathways. Phosphorylates SGK1 at Ser-78 and this is required for growth factor-induced cell cycle progression. Involved in the regulation of p53/TP53 by disrupting the PML-MDM2 interaction.
Dually phosphorylated on Thr-219 and Tyr-221, which activates the enzyme (By similarity). Autophosphorylated in vitro on threonine and tyrosine residues when the C-terminal part of the kinase, which could have a regulatory role, is absent.
Cytoplasm. Nucleus. Nucleus>PML body.
Note: Translocates to the nucleus upon activation.
Expressed in many adult tissues. Abundant in heart, placenta, lung, kidney and skeletal muscle. Not detectable in liver.
The second proline-rich region may interact with actin targeting the kinase to a specific location in the cell.
The TXY motif contains the threonine and tyrosine residues whose phosphorylation activates the MAP kinases.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. MAP kinase subfamily.
Research Fields
· Cellular Processes > Cellular community - eukaryotes > Gap junction. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Organismal Systems > Immune system > IL-17 signaling pathway. (View pathway)
· Organismal Systems > Nervous system > Neurotrophin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Oxytocin signaling pathway.
References
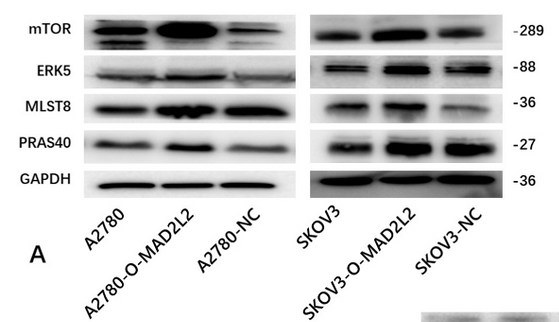
Application: WB Species: Human Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.