NGFR Antibody - #DF6821
| Product: | NGFR Antibody |
| Catalog: | DF6821 |
| Description: | Rabbit polyclonal antibody to NGFR |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Xenopus |
| Mol.Wt.: | 45kDa; 45kD(Calculated). |
| Uniprot: | P08138 |
| RRID: | AB_2838781 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF6821, RRID:AB_2838781.
Fold/Unfold
CD271; CD271 antigen; Gp80 LNGFR; Gp80-LNGFR; Low affinity nerve growth factor receptor; Low affinity neurotrophin receptor p75NTR; Low-affinity nerve growth factor receptor; Nerve growth factor receptor; Nerve growth factor receptor TNFR superfamily member 16; NGF receptor; Ngfr; p75 ICD; p75 Neurotrophin receptor; p75 NTR; p75(NTR); p75NTR; TNFR Superfamily Member 16; TNFRSF16; TNR16_HUMAN; Tumor necrosis factor receptor superfamily member 16;
Immunogens
A synthesized peptide derived from human NGFR, corresponding to a region within the internal amino acids.
- P08138 TNR16_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGAGATGRAMDGPRLLLLLLLGVSLGGAKEACPTGLYTHSGECCKACNLGEGVAQPCGANQTVCEPCLDSVTFSDVVSATEPCKPCTECVGLQSMSAPCVEADDAVCRCAYGYYQDETTGRCEACRVCEAGSGLVFSCQDKQNTVCEECPDGTYSDEANHVDPCLPCTVCEDTERQLRECTRWADAECEEIPGRWITRSTPPEGSDSTAPSTQEPEAPPEQDLIASTVAGVVTTVMGSSQPVVTRGTTDNLIPVYCSILAAVVVGLVAYIAFKRWNSCKQNKQGANSRPVNQTPPPEGEKLHSDSGISVDSQSLHDQQPHTQTASGQALKGDGGLYSSLPPAKREEVEKLLNGSAGDTWRHLAGELGYQPEHIDSFTHEACPVRALLASWATQDSATLDALLAALRRIQRADLVESLCSESTATSPV
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Low affinity receptor which can bind to NGF, BDNF, NTF3, and NTF4. Forms a heterodimeric receptor with SORCS2 that binds the precursor forms of NGF, BDNF and NTF3 with high affinity, and has much lower affinity for mature NGF and BDNF. Plays an important role in differentiation and survival of specific neuronal populations during development (By similarity). Can mediate cell survival as well as cell death of neural cells. Plays a role in the inactivation of RHOA. Plays a role in the regulation of the translocation of GLUT4 to the cell surface in adipocytes and skeletal muscle cells in response to insulin, probably by regulating RAB31 activity, and thereby contributes to the regulation of insulin-dependent glucose uptake (By similarity). Necessary for the circadian oscillation of the clock genes ARNTL/BMAL1, PER1, PER2 and NR1D1 in the suprachiasmatic nucleus (SCmgetaN) of the brain and in liver and of the genes involved in glucose and lipid metabolism in the liver.
N- and O-glycosylated.
O-linked glycans consist of Gal(1-3)GalNAc core elongated by 1 or 2 NeuNAc.
Phosphorylated on serine residues.
Cell membrane>Single-pass type I membrane protein. Perikaryon. Cell projection>Growth cone. Cell projection>Dendritic spine.
The death domain mediates interaction with RANBP9 (By similarity). It also mediates interaction with ARHGDIA and RIPK2 (PubMed:26646181).
The extracellular domain is responsible for interaction with NTRK1.
Research Fields
· Cellular Processes > Cell growth and death > Apoptosis - multiple species. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Ras signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Rap1 signaling pathway. (View pathway)
· Environmental Information Processing > Signaling molecules and interaction > Cytokine-cytokine receptor interaction. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
· Organismal Systems > Nervous system > Neurotrophin signaling pathway. (View pathway)
References
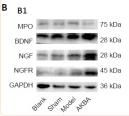
Application: WB Species: Rat Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.