BTRC Antibody - #DF6534
| Product: | BTRC Antibody |
| Catalog: | DF6534 |
| Description: | Rabbit polyclonal antibody to BTRC |
| Application: | WB IHC IF/ICC |
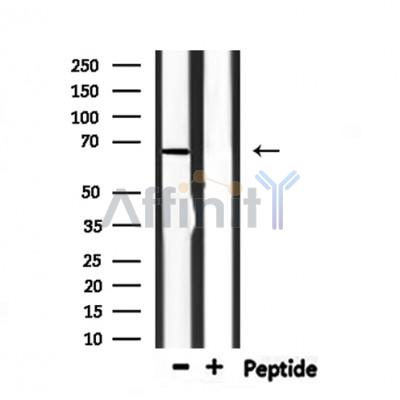
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 69kDa; 69kD(Calculated). |
| Uniprot: | Q9Y297 |
| RRID: | AB_2838496 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF6534, RRID:AB_2838496.
Fold/Unfold
Beta transducin repeat containing E3 ubiquitin protein ligase; beta TrCP1; beta-transducin repeat containing; BETA-TRCP; betaTrCP; btrC; bTrCP; bTrCP1; E3RSIkappaB; F box and WD repeat protein 1B; F box and WD repeats protein beta TrCP; F-box and WD repeats protein beta-TrCP; F-box/WD repeat protein 1A; F-box/WD repeat-containing protein 1A; FBW1A; FBW1A_HUMAN; FBXW1; FBXW1A; FWD1; HOS; mKIAA4123; pIkappaBalpha-E3 receptor subunit; Slimb;
Immunogens
A synthesized peptide derived from human BTRC, corresponding to a region within N-terminal amino acids.
- Q9Y297 FBW1A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MDPAEAVLQEKALKFMCSMPRSLWLGCSSLADSMPSLRCLYNPGTGALTAFQNSSEREDCNNGEPPRKIIPEKNSLRQTYNSCARLCLNQETVCLASTAMKTENCVAKTKLANGTSSMIVPKQRKLSASYEKEKELCVKYFEQWSESDQVEFVEHLISQMCHYQHGHINSYLKPMLQRDFITALPARGLDHIAENILSYLDAKSLCAAELVCKEWYRVTSDGMLWKKLIERMVRTDSLWRGLAERRGWGQYLFKNKPPDGNAPPNSFYRALYPKIIQDIETIESNWRCGRHSLQRIHCRSETSKGVYCLQYDDQKIVSGLRDNTIKIWDKNTLECKRILTGHTGSVLCLQYDERVIITGSSDSTVRVWDVNTGEMLNTLIHHCEAVLHLRFNNGMMVTCSKDRSIAVWDMASPTDITLRRVLVGHRAAVNVVDFDDKYIVSASGDRTIKVWNTSTCEFVRTLNGHKRGIACLQYRDRLVVSGSSDNTIRLWDIECGACLRVLEGHEELVRCIRFDNKRIVSGAYDGKIKVWDLVAALDPRAPAGTLCLRTLVEHSGRVFRLQFDEFQIVSSSHDDTILIWDFLNDPAAQAEPPRSPSRTYTYISR
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Substrate recognition component of a SCF (SKP1-CUL1-F-box protein) E3 ubiquitin-protein ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of target proteins. Recognizes and binds to phosphorylated target proteins. SCF(BTRC) mediates the ubiquitination of CTNNB1 and participates in Wnt signaling. SCF(BTRC) mediates the ubiquitination of phosphorylated NFKB1, ATF4, CDC25A, DLG1, FBXO5, PER1, SMAD3, SMAD4, SNAI1 and probably NFKB2. SCF(BTRC) mediates the ubiquitination of NFKBIA, NFKBIB and NFKBIE; the degradation frees the associated NFKB1 to translocate into the nucleus and to activate transcription. Ubiquitination of NFKBIA occurs at 'Lys-21' and 'Lys-22'. SCF(BTRC) mediates the ubiquitination of CEP68; this is required for centriole separation during mitosis. SCF(BTRC) mediates the ubiquitination and subsequent degradation of nuclear NFE2L1 (By similarity). Has an essential role in the control of the clock-dependent transcription via degradation of phosphorylated PER1 and PER2. May be involved in ubiquitination and subsequent proteasomal degradation through a DBB1-CUL4 E3 ubiquitin-protein ligase. Required for activation of NFKB-mediated transcription by IL1B, MAP3K14, MAP3K1, IKBKB and TNF. Required for proteolytic processing of GLI3. Mediates ubiquitination of REST, thereby leading to its proteasomal degradation.
Cytoplasm. Nucleus.
Expressed in epididymis (at protein level).
The N-terminal D domain mediates homodimerization.
Research Fields
· Cellular Processes > Cell growth and death > Oocyte meiosis. (View pathway)
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Environmental Information Processing > Signal transduction > Wnt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hedgehog signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hippo signaling pathway. (View pathway)
· Genetic Information Processing > Folding, sorting and degradation > Ubiquitin mediated proteolysis. (View pathway)
· Human Diseases > Infectious diseases: Bacterial > Shigellosis.
· Organismal Systems > Environmental adaptation > Circadian rhythm. (View pathway)
References
Application: WB Species: human Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.