alpha 2 Macroglobulin/A2M Antibody - #DF6469
| Product: | alpha 2 Macroglobulin/A2M Antibody |
| Catalog: | DF6469 |
| Description: | Rabbit polyclonal antibody to alpha 2 Macroglobulin/A2M |
| Application: | WB IHC |
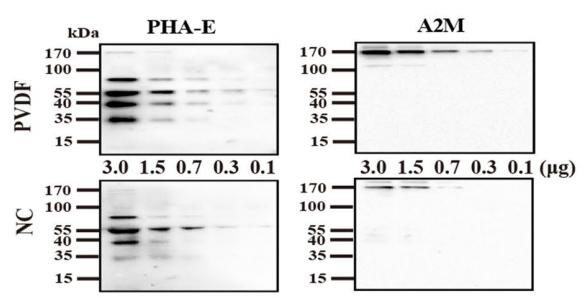
| Cited expt.: | WB |
| Reactivity: | Human, Mouse |
| Mol.Wt.: | 163kDa; 163kD(Calculated). |
| Uniprot: | P01023 |
| RRID: | AB_2838431 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF6469, RRID:AB_2838431.
Fold/Unfold
A2m; A2MG_HUMAN; Alpha 2 M; Alpha 2M; Alpha-2-M; Alpha-2-macroglobulin; C3 and PZP-like alpha-2-macroglobulin domain-containing protein 5; CPAMD5; DKFZp779B086; FWP007; S863 7;
Immunogens
A synthesized peptide derived from human alpha 2 Macroglobulin/A2M, corresponding to a region within the internal amino acids.
- P01023 A2MG_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGKNKLLHPSLVLLLLVLLPTDASVSGKPQYMVLVPSLLHTETTEKGCVLLSYLNETVTVSASLESVRGNRSLFTDLEAENDVLHCVAFAVPKSSSNEEVMFLTVQVKGPTQEFKKRTTVMVKNEDSLVFVQTDKSIYKPGQTVKFRVVSMDENFHPLNELIPLVYIQDPKGNRIAQWQSFQLEGGLKQFSFPLSSEPFQGSYKVVVQKKSGGRTEHPFTVEEFVLPKFEVQVTVPKIITILEEEMNVSVCGLYTYGKPVPGHVTVSICRKYSDASDCHGEDSQAFCEKFSGQLNSHGCFYQQVKTKVFQLKRKEYEMKLHTEAQIQEEGTVVELTGRQSSEITRTITKLSFVKVDSHFRQGIPFFGQVRLVDGKGVPIPNKVIFIRGNEANYYSNATTDEHGLVQFSINTTNVMGTSLTVRVNYKDRSPCYGYQWVSEEHEEAHHTAYLVFSPSKSFVHLEPMSHELPCGHTQTVQAHYILNGGTLLGLKKLSFYYLIMAKGGIVRTGTHGLLVKQEDMKGHFSISIPVKSDIAPVARLLIYAVLPTGDVIGDSAKYDVENCLANKVDLSFSPSQSLPASHAHLRVTAAPQSVCALRAVDQSVLLMKPDAELSASSVYNLLPEKDLTGFPGPLNDQDNEDCINRHNVYINGITYTPVSSTNEKDMYSFLEDMGLKAFTNSKIRKPKMCPQLQQYEMHGPEGLRVGFYESDVMGRGHARLVHVEEPHTETVRKYFPETWIWDLVVVNSAGVAEVGVTVPDTITEWKAGAFCLSEDAGLGISSTASLRAFQPFFVELTMPYSVIRGEAFTLKATVLNYLPKCIRVSVQLEASPAFLAVPVEKEQAPHCICANGRQTVSWAVTPKSLGNVNFTVSAEALESQELCGTEVPSVPEHGRKDTVIKPLLVEPEGLEKETTFNSLLCPSGGEVSEELSLKLPPNVVEESARASVSVLGDILGSAMQNTQNLLQMPYGCGEQNMVLFAPNIYVLDYLNETQQLTPEIKSKAIGYLNTGYQRQLNYKHYDGSYSTFGERYGRNQGNTWLTAFVLKTFAQARAYIFIDEAHITQALIWLSQRQKDNGCFRSSGSLLNNAIKGGVEDEVTLSAYITIALLEIPLTVTHPVVRNALFCLESAWKTAQEGDHGSHVYTKALLAYAFALAGNQDKRKEVLKSLNEEAVKKDNSVHWERPQKPKAPVGHFYEPQAPSAEVEMTSYVLLAYLTAQPAPTSEDLTSATNIVKWITKQQNAQGGFSSTQDTVVALHALSKYGAATFTRTGKAAQVTIQSSGTFSSKFQVDNNNRLLLQQVSLPELPGEYSMKVTGEGCVYLQTSLKYNILPEKEEFPFALGVQTLPQTCDEPKAHTSFQISLSVSYTGSRSASNMAIVDVKMVSGFIPLKPTVKMLERSNHVSRTEVSSNHVLIYLDKVSNQTLSLFFTVLQDVPVRDLKPAIVKVYDYYETDEFAIAEYNAPCSKDLGNA
Research Backgrounds
Is able to inhibit all four classes of proteinases by a unique 'trapping' mechanism. This protein has a peptide stretch, called the 'bait region' which contains specific cleavage sites for different proteinases. When a proteinase cleaves the bait region, a conformational change is induced in the protein which traps the proteinase. The entrapped enzyme remains active against low molecular weight substrates (activity against high molecular weight substrates is greatly reduced). Following cleavage in the bait region, a thioester bond is hydrolyzed and mediates the covalent binding of the protein to the proteinase.
Secreted.
Secreted in plasma.
Belongs to the protease inhibitor I39 (alpha-2-macroglobulin) family.
Research Fields
· Organismal Systems > Immune system > Complement and coagulation cascades. (View pathway)
References
Application: WB Species: Human Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.