Annexin A2 Antibody - #DF6468
| Product: | Annexin A2 Antibody |
| Catalog: | DF6468 |
| Description: | Rabbit polyclonal antibody to Annexin A2 |
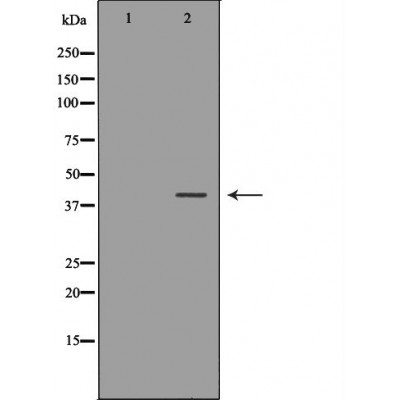
| Application: | WB IHC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken |
| Mol.Wt.: | 39kDa; 39kD(Calculated). |
| Uniprot: | P07355 |
| RRID: | AB_2838430 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF6468, RRID:AB_2838430.
Fold/Unfold
Annexin A2; Annexin II; Annexin II, heavy chain; Annexin-2; ANX 2; ANX2; ANX2L4; ANXA2; ANXA2_HUMAN; arylsulfatase B; CAL1H; Calpactin I heavy chain; calpactin I heavy polypeptide (p36); Calpactin I heavy polypeptide; Calpactin-1 heavy chain; chromobindin 8; Chromobindin-8; Epididymis secretory protein Li 270; HEL S 270; LIP2; Lipocortin II; LPC2; LPC2D; p36; P36 protein; PAP-IV; Placental anticoagulant protein IV; Protein I;
Immunogens
A synthesized peptide derived from human Annexin A2, corresponding to a region within the internal amino acids.
- P07355 ANXA2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSTVHEILCKLSLEGDHSTPPSAYGSVKAYTNFDAERDALNIETAIKTKGVDEVTIVNILTNRSNAQRQDIAFAYQRRTKKELASALKSALSGHLETVILGLLKTPAQYDASELKASMKGLGTDEDSLIEIICSRTNQELQEINRVYKEMYKTDLEKDIISDTSGDFRKLMVALAKGRRAEDGSVIDYELIDQDARDLYDAGVKRKGTDVPKWISIMTERSVPHLQKVFDRYKSYSPYDMLESIRKEVKGDLENAFLNLVQCIQNKPLYFADRLYDSMKGKGTRDKVLIRIMVSRSEVDMLKIRSEFKRKYGKSLYYYIQQDTKGDYQKALLYLCGGDD
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Calcium-regulated membrane-binding protein whose affinity for calcium is greatly enhanced by anionic phospholipids. It binds two calcium ions with high affinity. May be involved in heat-stress response. Inhibits PCSK9-enhanced LDLR degradation, probably reduces PCSK9 protein levels via a translational mechanism but also competes with LDLR for binding with PCSK9.
Phosphorylation of Tyr-24 enhances heat stress-induced translocation to the cell surface.
ISGylated.
Secreted>Extracellular space>Extracellular matrix>Basement membrane. Melanosome.
Note: In the lamina beneath the plasma membrane. Identified by mass spectrometry in melanosome fractions from stage I to stage IV. Translocated from the cytoplasm to the cell surface through a Golgi-independent mechanism.
A pair of annexin repeats may form one binding site for calcium and phospholipid.
Belongs to the annexin family.
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.