EHMT2 Antibody - #DF6379
| Product: | EHMT2 Antibody |
| Catalog: | DF6379 |
| Description: | Rabbit polyclonal antibody to EHMT2 |
| Application: | WB IHC |
| Cited expt.: | |
| Reactivity: | Human, Mouse, Rat |
| Mol.Wt.: | 132kDa; 132kD(Calculated). |
| Uniprot: | Q96KQ7 |
| RRID: | AB_2838342 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF6379, RRID:AB_2838342.
Fold/Unfold
Ankyrin repeat containing protein; Bat 8; Bat8; C6orf30; DKFZp686H08213; EHMT 2; Ehmt2; EHMT2_HUMAN; Euchromatic histone lysine methyltransferase 2; Euchromatic histone lysine N methyltransferase 2; Euchromatic histone-lysine N-methyltransferase 2; FLJ35547; G 9a; G9 a; G9a protein; G9A; G9A histone methyltransferase; GAT8; H3 K9 HMTase 3; H3-K9-HMTase 3; Histone H3 K9 methyltransferase 3; Histone H3 K9 methyltransferase3; Histone H3-K9 methyltransferase 3; Histone lysine N methyltransferase; Histone lysine N methyltransferase EHMT2; Histone lysine N methyltransferase, H3 lysine 9 specific 3; Histone lysine N methyltransferase, H3 lysine 9 specific3; Histone-lysine N-methyltransferase EHMT2; HLA B associated transcript 8; HLA-B-associated transcript 8; KMT 1C; KMT1 C; Lysine N methyltransferase 1C; Lysine N-methyltransferase 1C; NG 36; NG36; Protein G9a;
Immunogens
A synthesized peptide derived from human EHMT2, corresponding to a region within N-terminal amino acids.
Expressed in all tissues examined, with high levels in fetal liver, thymus, lymph node, spleen and peripheral blood leukocytes and lower level in bone marrow.
- Q96KQ7 EHMT2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAAAAGAAAAAAAEGEAPAEMGALLLEKETRGATERVHGSLGDTPRSEETLPKATPDSLEPAGPSSPASVTVTVGDEGADTPVGATPLIGDESENLEGDGDLRGGRILLGHATKSFPSSPSKGGSCPSRAKMSMTGAGKSPPSVQSLAMRLLSMPGAQGAAAAGSEPPPATTSPEGQPKVHRARKTMSKPGNGQPPVPEKRPPEIQHFRMSDDVHSLGKVTSDLAKRRKLNSGGGLSEELGSARRSGEVTLTKGDPGSLEEWETVVGDDFSLYYDSYSVDERVDSDSKSEVEALTEQLSEEEEEEEEEEEEEEEEEEEEEEEEDEESGNQSDRSGSSGRRKAKKKWRKDSPWVKPSRKRRKREPPRAKEPRGVNGVGSSGPSEYMEVPLGSLELPSEGTLSPNHAGVSNDTSSLETERGFEELPLCSCRMEAPKIDRISERAGHKCMATESVDGELSGCNAAILKRETMRPSSRVALMVLCETHRARMVKHHCCPGCGYFCTAGTFLECHPDFRVAHRFHKACVSQLNGMVFCPHCGEDASEAQEVTIPRGDGVTPPAGTAAPAPPPLSQDVPGRADTSQPSARMRGHGEPRRPPCDPLADTIDSSGPSLTLPNGGCLSAVGLPLGPGREALEKALVIQESERRKKLRFHPRQLYLSVKQGELQKVILMLLDNLDPNFQSDQQSKRTPLHAAAQKGSVEICHVLLQAGANINAVDKQQRTPLMEAVVNNHLEVARYMVQRGGCVYSKEEDGSTCLHHAAKIGNLEMVSLLLSTGQVDVNAQDSGGWTPIIWAAEHKHIEVIRMLLTRGADVTLTDNEENICLHWASFTGSAAIAEVLLNARCDLHAVNYHGDTPLHIAARESYHDCVLLFLSRGANPELRNKEGDTAWDLTPERSDVWFALQLNRKLRLGVGNRAIRTEKIICRDVARGYENVPIPCVNGVDGEPCPEDYKYISENCETSTMNIDRNITHLQHCTCVDDCSSSNCLCGQLSIRCWYDKDGRLLQEFNKIEPPLIFECNQACSCWRNCKNRVVQSGIKVRLQLYRTAKMGWGVRALQTIPQGTFICEYVGELISDAEADVREDDSYLFDLDNKDGEVYCIDARYYGNISRFINHLCDPNIIPVRVFMLHQDLRFPRIAFFSSRDIRTGEELGFDYGDRFWDIKSKYFTCQCGSEKCKHSAEAIALEQSRLARLDPHPELLPELGSLPPVNT
Research Backgrounds
Histone methyltransferase that specifically mono- and dimethylates 'Lys-9' of histone H3 (H3K9me1 and H3K9me2, respectively) in euchromatin. H3K9me represents a specific tag for epigenetic transcriptional repression by recruiting HP1 proteins to methylated histones. Also mediates monomethylation of 'Lys-56' of histone H3 (H3K56me1) in G1 phase, leading to promote interaction between histone H3 and PCNA and regulating DNA replication. Also weakly methylates 'Lys-27' of histone H3 (H3K27me). Also required for DNA methylation, the histone methyltransferase activity is not required for DNA methylation, suggesting that these 2 activities function independently. Probably targeted to histone H3 by different DNA-binding proteins like E2F6, MGA, MAX and/or DP1. May also methylate histone H1. In addition to the histone methyltransferase activity, also methylates non-histone proteins: mediates dimethylation of 'Lys-373' of p53/TP53. Also methylates CDYL, WIZ, ACIN1, DNMT1, HDAC1, ERCC6, KLF12 and itself.
Methylated at Lys-185; automethylated.
Nucleus. Chromosome.
Note: Associates with euchromatic regions. Does not associate with heterochromatin.
Expressed in all tissues examined, with high levels in fetal liver, thymus, lymph node, spleen and peripheral blood leukocytes and lower level in bone marrow.
The SET domain mediates interaction with WIZ.
The ANK repeats bind H3K9me1 and H3K9me2.
In the pre-SET domain, Cys residues bind 3 zinc ions that are arranged in a triangular cluster; some of these Cys residues contribute to the binding of two zinc ions within the cluster.
Belongs to the class V-like SAM-binding methyltransferase superfamily. Histone-lysine methyltransferase family. Suvar3-9 subfamily.
Research Fields
· Metabolism > Amino acid metabolism > Lysine degradation.
· Organismal Systems > Aging > Longevity regulating pathway. (View pathway)
References
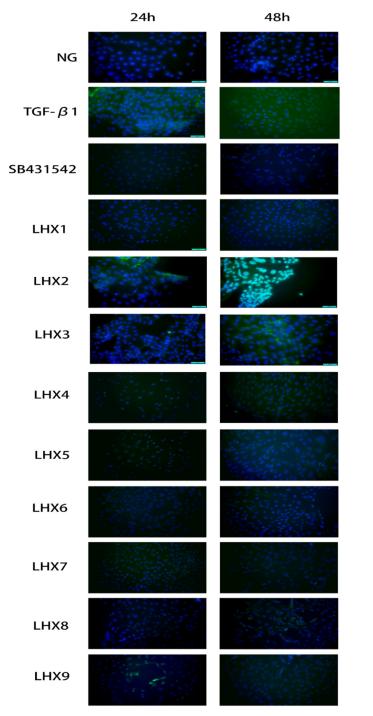
Application: IF/ICC Species: Mouse Sample: MTEC cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.