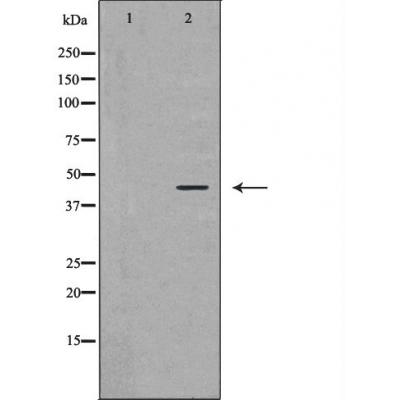
Ataxin 3 Antibody - #DF6375
| Product: | Ataxin 3 Antibody |
| Catalog: | DF6375 |
| Description: | Rabbit polyclonal antibody to Ataxin 3 |
| Application: | WB IHC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 43kDa; 41kD(Calculated). |
| Uniprot: | P54252 |
| RRID: | AB_2838339 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF6375, RRID:AB_2838339.
Fold/Unfold
AT3; Ataxin 3; ataxin 3 variant h; ataxin 3 variant m; ataxin 3 variant ref; Ataxin-3; ATX3; ATX3_HUMAN; ATXN3; EC 3.4.22.; JOS; Josephin; Machado Joseph disease (spinocerebellar ataxia 3, olivopontocerebellar ataxia 3, autosomal dominant, ataxin 3); Machado Joseph disease; Machado Joseph disease protein 1; Machado-Joseph disease protein 1; Machado-Joseph disease protein 1 homolog; MJD; MJD gene; MJD1; Olivopontocerebellar ataxia 3; OTTHUMP00000221583; OTTHUMP00000221585; OTTHUMP00000221586; OTTHUMP00000221587; OTTHUMP00000231995; OTTHUMP00000231997; Rsca3; SCA3; SCA3 gene; Spinocerebellar ataxia type 3 protein;
Immunogens
A synthesized peptide derived from human Ataxin 3, corresponding to a region within the internal amino acids.
- P54252 ATX3_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MESIFHEKQEGSLCAQHCLNNLLQGEYFSPVELSSIAHQLDEEERMRMAEGGVTSEDYRTFLQQPSGNMDDSGFFSIQVISNALKVWGLELILFNSPEYQRLRIDPINERSFICNYKEHWFTVRKLGKQWFNLNSLLTGPELISDTYLALFLAQLQQEGYSIFVVKGDLPDCEADQLLQMIRVQQMHRPKLIGEELAQLKEQRVHKTDLERVLEANDGSGMLDEDEEDLQRALALSRQEIDMEDEEADLRRAIQLSMQGSSRNISQDMTQTSGTNLTSEELRKRREAYFEKQQQKQQQQQQQQQQGDLSGQSSHPCERPATSSGALGSDLGDAMSEEDMLQAAVTMSLETVRNDLKTEGKK
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Deubiquitinating enzyme involved in protein homeostasis maintenance, transcription, cytoskeleton regulation, myogenesis and degradation of misfolded chaperone substrates. Binds long polyubiquitin chains and trims them, while it has weak or no activity against chains of 4 or less ubiquitins. Involved in degradation of misfolded chaperone substrates via its interaction with STUB1/CHIP: recruited to monoubiquitinated STUB1/CHIP, and restricts the length of ubiquitin chain attached to STUB1/CHIP substrates and preventing further chain extension (By similarity). Interacts with key regulators of transcription and represses transcription: acts as a histone-binding protein that regulates transcription. Regulates autophagy via the deubiquitination of 'Lys-402' of BECN1 leading to the stabilization of BECN1.
Monoubiquitinated N-terminally by UBE2W, possibly leading to activate the deubiquitinating enzyme activity.
Nucleus matrix. Nucleus.
Note: Predominantly nuclear, but not exclusively, inner nuclear matrix.
Ubiquitous.
The UIM domains bind ubiquitin and interact with various E3 ubiquitin-protein ligase, such as STUB1/CHIP. They are essential to limit the length of ubiquitin chains (By similarity).
The poly-Gln domain is involved in the interaction with BECN1 and subsequent starvation-induced autophagy (PubMed:28445460).
Research Fields
· Genetic Information Processing > Folding, sorting and degradation > Protein processing in endoplasmic reticulum. (View pathway)
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.