SGK1 Antibody - #DF6188
| Product: | SGK1 Antibody |
| Catalog: | DF6188 |
| Description: | Rabbit polyclonal antibody to SGK1 |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Chicken |
| Mol.Wt.: | 49kDa; 49kD(Calculated). |
| Uniprot: | O00141 |
| RRID: | AB_2838154 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF6188, RRID:AB_2838154.
Fold/Unfold
OTTHUMP00000017247; Serine/threonine protein kinase SGK; Serine/threonine protein kinase Sgk1; Serine/threonine-protein kinase Sgk1; Serum and glucocorticoid regulated kinase; Serum/glucocorticoid regulated kinase 1; Serum/glucocorticoid regulated kinase; Serum/glucocorticoid-regulated kinase 1; SGK 1; SGK; SGK1; Sgk1 variant i3; SGK1_HUMAN;
Immunogens
A synthesized peptide derived from human SGK1, corresponding to a region within the internal amino acids.
Expressed in most tissues with highest levels in the pancreas, followed by placenta, kidney and lung. Isoform 2 is strongly expressed in brain and pancreas, weaker in heart, placenta, lung, liver and skeletal muscle.
- O00141 SGK1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MTVKTEAAKGTLTYSRMRGMVAILIAFMKQRRMGLNDFIQKIANNSYACKHPEVQSILKISQPQEPELMNANPSPPPSPSQQINLGPSSNPHAKPSDFHFLKVIGKGSFGKVLLARHKAEEVFYAVKVLQKKAILKKKEEKHIMSERNVLLKNVKHPFLVGLHFSFQTADKLYFVLDYINGGELFYHLQRERCFLEPRARFYAAEIASALGYLHSLNIVYRDLKPENILLDSQGHIVLTDFGLCKENIEHNSTTSTFCGTPEYLAPEVLHKQPYDRTVDWWCLGAVLYEMLYGLPPFYSRNTAEMYDNILNKPLQLKPNITNSARHLLEGLLQKDRTKRLGAKDDFMEIKSHVFFSLINWDDLINKKITPPFNPNVSGPNDLRHFDPEFTEEPVPNSIGKSPDSVLVTASVKEAAEAFLGFSYAPPTDSFL
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Serine/threonine-protein kinase which is involved in the regulation of a wide variety of ion channels, membrane transporters, cellular enzymes, transcription factors, neuronal excitability, cell growth, proliferation, survival, migration and apoptosis. Plays an important role in cellular stress response. Contributes to regulation of renal Na(+) retention, renal K(+) elimination, salt appetite, gastric acid secretion, intestinal Na(+)/H(+) exchange and nutrient transport, insulin-dependent salt sensitivity of blood pressure, salt sensitivity of peripheral glucose uptake, cardiac repolarization and memory consolidation. Up-regulates Na(+) channels: SCNN1A/ENAC, SCN5A and ASIC1/ACCN2, K(+) channels: KCNJ1/ROMK1, KCNA1-5, KCNQ1-5 and KCNE1, epithelial Ca(2+) channels: TRPV5 and TRPV6, chloride channels: BSND, CLCN2 and CFTR, glutamate transporters: SLC1A3/EAAT1, SLC1A2 /EAAT2, SLC1A1/EAAT3, SLC1A6/EAAT4 and SLC1A7/EAAT5, amino acid transporters: SLC1A5/ASCT2, SLC38A1/SN1 and SLC6A19, creatine transporter: SLC6A8, Na(+)/dicarboxylate cotransporter: SLC13A2/NADC1, Na(+)-dependent phosphate cotransporter: SLC34A2/NAPI-2B, glutamate receptor: GRIK2/GLUR6. Up-regulates carriers: SLC9A3/NHE3, SLC12A1/NKCC2, SLC12A3/NCC, SLC5A3/SMIT, SLC2A1/GLUT1, SLC5A1/SGLT1 and SLC15A2/PEPT2. Regulates enzymes: GSK3A/B, PMM2 and Na(+)/K(+) ATPase, and transcription factors: CTNNB1 and nuclear factor NF-kappa-B. Stimulates sodium transport into epithelial cells by enhancing the stability and expression of SCNN1A/ENAC. This is achieved by phosphorylating the NEDD4L ubiquitin E3 ligase, promoting its interaction with 14-3-3 proteins, thereby preventing it from binding to SCNN1A/ENAC and targeting it for degradation. Regulates store-operated Ca(+2) entry (SOCE) by stimulating ORAI1 and STIM1. Regulates KCNJ1/ROMK1 directly via its phosphorylation or indirectly via increased interaction with SLC9A3R2/NHERF2. Phosphorylates MDM2 and activates MDM2-dependent ubiquitination of p53/TP53. Phosphorylates MAPT/TAU and mediates microtubule depolymerization and neurite formation in hippocampal neurons. Phosphorylates SLC2A4/GLUT4 and up-regulates its activity. Phosphorylates APBB1/FE65 and promotes its localization to the nucleus. Phosphorylates MAPK1/ERK2 and activates it by enhancing its interaction with MAP2K1/MEK1 and MAP2K2/MEK2. Phosphorylates FBXW7 and plays an inhibitory role in the NOTCH1 signaling. Phosphorylates FOXO1 resulting in its relocalization from the nucleus to the cytoplasm. Phosphorylates FOXO3, promoting its exit from the nucleus and interference with FOXO3-dependent transcription. Phosphorylates BRAF and MAP3K3/MEKK3 and inhibits their activity. Phosphorylates SLC9A3/NHE3 in response to dexamethasone, resulting in its activation and increased localization at the cell membrane. Phosphorylates CREB1. Necessary for vascular remodeling during angiogenesis. Sustained high levels and activity may contribute to conditions such as hypertension and diabetic nephropathy. Isoform 2 exhibited a greater effect on cell plasma membrane expression of SCNN1A/ENAC and Na(+) transport than isoform 1.
Regulated by phosphorylation. Activated by phosphorylation on Ser-422 by mTORC2, transforming it into a substrate for PDPK1 which phosphorylates it on Thr-256. Phosphorylation on Ser-397 and Ser-401 are also essential for its activity. Phosphorylation on Ser-78 by MAPK7 is required for growth factor-induced cell cycle progression.
Ubiquitinated by NEDD4L; which promotes proteasomal degradation. Ubiquitinated by SYVN1 at the endoplasmic reticulum; which promotes rapid proteasomal degradation and maintains a high turnover rate in resting cells. Isoform 2 shows enhanced stability.
Cytoplasm. Nucleus. Endoplasmic reticulum membrane. Cell membrane. Mitochondrion.
Note: The subcellular localization is controlled by the cell cycle, as well as by exposure to specific hormones and environmental stress stimuli. In proliferating cells, it shuttles between the nucleus and cytoplasm in synchrony with the cell cycle, and in serum/growth factor-stimulated cells it resides in the nucleus. In contrast, after exposure to environmental stress or treatment with glucocorticoids, it is detected in the cytoplasm and with certain stress conditions is associated with the mitochondria. In osmoregulation through the epithelial sodium channel, it can be localized to the cytoplasmic surface of the cell membrane. Nuclear, upon phosphorylation.
Cell membrane.
Expressed in most tissues with highest levels in the pancreas, followed by placenta, kidney and lung. Isoform 2 is strongly expressed in brain and pancreas, weaker in heart, placenta, lung, liver and skeletal muscle.
Isoform 2 subcellular localization at the cell membrane and resistance to proteasomal degradation is mediated by the sequences within the first 120 amino acids.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family.
Research Fields
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > mTOR signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Organismal Systems > Excretory system > Aldosterone-regulated sodium reabsorption.
References
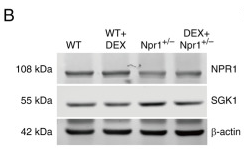
Application: WB Species: Mouse Sample: heart tissue
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.