PSME3 Antibody - #DF6074
| Product: | PSME3 Antibody |
| Catalog: | DF6074 |
| Description: | Rabbit polyclonal antibody to PSME3 |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 30kDa; 30kD(Calculated). |
| Uniprot: | P61289 |
| RRID: | AB_2838042 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF6074, RRID:AB_2838042.
Fold/Unfold
11S regulator complex gamma subunit; 11S regulator complex subunit gamma; Activator of multicatalytic protease subunit 3; Ki; Ki antigen; Ki nuclear autoantigen; Ki, PA28 gamma; PA28 gamma; PA28g; PA28gamma; Proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki); Proteasome (prosome, macropain) activator subunit 3; Proteasome activator 28 gamma; Proteasome activator 28 subunit gamma; Proteasome activator complex subunit 3; Proteasome activator subunit 3; PSME3; PSME3_HUMAN; REG GAMMA; REG-gamma;
Immunogens
A synthesized peptide derived from human PSME3, corresponding to a region within N-terminal amino acids.
- P61289 PSME3_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MASLLKVDQEVKLKVDSFRERITSEAEDLVANFFPKKLLELDSFLKEPILNIHDLTQIHSDMNLPVPDPILLTNSHDGLDGPTYKKRRLDECEEAFQGTKVFVMPNGMLKSNQQLVDIIEKVKPEIRLLIEKCNTVKMWVQLLIPRIEDGNNFGVSIQEETVAELRTVESEAASYLDQISRYYITRAKLVSKIAKYPHVEDYRRTVTEIDEKEYISLRLIISELRNQYVTLHDMILKNIEKIKRPRSSNAETLY
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Subunit of the 11S REG-gamma (also called PA28-gamma) proteasome regulator, a doughnut-shaped homoheptamer which associates with the proteasome. 11S REG-gamma activates the trypsin-like catalytic subunit of the proteasome but inhibits the chymotrypsin-like and postglutamyl-preferring (PGPH) subunits. Facilitates the MDM2-p53/TP53 interaction which promotes ubiquitination- and MDM2-dependent proteasomal degradation of p53/TP53, limiting its accumulation and resulting in inhibited apoptosis after DNA damage. May also be involved in cell cycle regulation. Mediates CCAR2 and CHEK2-dependent SIRT1 inhibition.
Phosphorylated by MAP3K3 (By similarity). Phosphorylation at Ser-247 promotes its association with CCAR2.
Acetylation at the major site Lys-195 is important for oligomerization and ability to degrade its target substrates. Deacetylated by SIRT1.
Nucleus. Cytoplasm.
Note: Localizes to the cytoplasm during mitosis following nuclear envelope breakdown at this distinct stage of the cell cycle which allows its interaction with MAP3K3 kinase.
The C-terminal sequences affect heptamer stability and proteasome affinity.
Belongs to the PA28 family.
Research Fields
· Genetic Information Processing > Folding, sorting and degradation > Proteasome.
· Human Diseases > Infectious diseases: Viral > Hepatitis C.
· Organismal Systems > Immune system > Antigen processing and presentation. (View pathway)
References
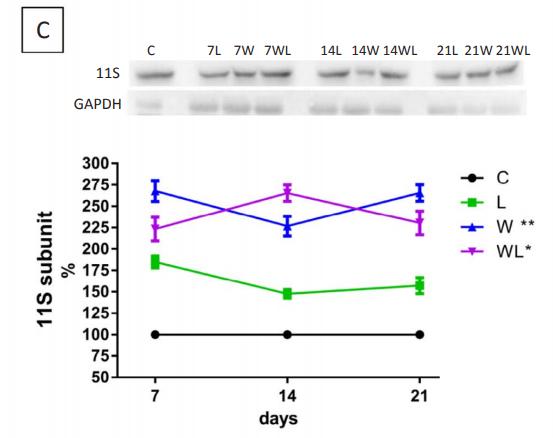
Application: WB Species: rat Sample: gastrocnemius muscle tissue
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.