TERT Antibody - #AF5413
| Product: | TERT Antibody |
| Catalog: | AF5413 |
| Description: | Rabbit polyclonal antibody to TERT |
| Application: | WB |
| Cited expt.: | WB |
| Reactivity: | Human |
| Prediction: | Pig, Dog |
| Mol.Wt.: | 127 kDa; 127kD(Calculated). |
| Uniprot: | O14746 |
| RRID: | AB_2837897 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF5413, RRID:AB_2837897.
Fold/Unfold
CMM9; DKCA2; DKCB4; EST2; HEST2; htert; hTRT; PFBMFT1; TCS1; Telomerase associated protein 2; Telomerase catalytic subunit; Telomerase reverse transcriptase; Telomerase-associated protein 2; Telomere Reverse Transcriptase; TERT; TERT_HUMAN; TP2; TRT;
Immunogens
A synthesized peptide derived from human TERT, corresponding to a region within the internal amino acids.
Expressed at a high level in thymocyte subpopulations, at an intermediate level in tonsil T-lymphocytes, and at a low to undetectable level in peripheral blood T-lymphocytes.
- O14746 TERT_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MPRAPRCRAVRSLLRSHYREVLPLATFVRRLGPQGWRLVQRGDPAAFRALVAQCLVCVPWDARPPPAAPSFRQVSCLKELVARVLQRLCERGAKNVLAFGFALLDGARGGPPEAFTTSVRSYLPNTVTDALRGSGAWGLLLRRVGDDVLVHLLARCALFVLVAPSCAYQVCGPPLYQLGAATQARPPPHASGPRRRLGCERAWNHSVREAGVPLGLPAPGARRRGGSASRSLPLPKRPRRGAAPEPERTPVGQGSWAHPGRTRGPSDRGFCVVSPARPAEEATSLEGALSGTRHSHPSVGRQHHAGPPSTSRPPRPWDTPCPPVYAETKHFLYSSGDKEQLRPSFLLSSLRPSLTGARRLVETIFLGSRPWMPGTPRRLPRLPQRYWQMRPLFLELLGNHAQCPYGVLLKTHCPLRAAVTPAAGVCAREKPQGSVAAPEEEDTDPRRLVQLLRQHSSPWQVYGFVRACLRRLVPPGLWGSRHNERRFLRNTKKFISLGKHAKLSLQELTWKMSVRDCAWLRRSPGVGCVPAAEHRLREEILAKFLHWLMSVYVVELLRSFFYVTETTFQKNRLFFYRKSVWSKLQSIGIRQHLKRVQLRELSEAEVRQHREARPALLTSRLRFIPKPDGLRPIVNMDYVVGARTFRREKRAERLTSRVKALFSVLNYERARRPGLLGASVLGLDDIHRAWRTFVLRVRAQDPPPELYFVKVDVTGAYDTIPQDRLTEVIASIIKPQNTYCVRRYAVVQKAAHGHVRKAFKSHVSTLTDLQPYMRQFVAHLQETSPLRDAVVIEQSSSLNEASSGLFDVFLRFMCHHAVRIRGKSYVQCQGIPQGSILSTLLCSLCYGDMENKLFAGIRRDGLLLRLVDDFLLVTPHLTHAKTFLRTLVRGVPEYGCVVNLRKTVVNFPVEDEALGGTAFVQMPAHGLFPWCGLLLDTRTLEVQSDYSSYARTSIRASLTFNRGFKAGRNMRRKLFGVLRLKCHSLFLDLQVNSLQTVCTNIYKILLLQAYRFHACVLQLPFHQQVWKNPTFFLRVISDTASLCYSILKAKNAGMSLGAKGAAGPLPSEAVQWLCHQAFLLKLTRHRVTYVPLLGSLRTAQTQLSRKLPGTTLTALEAAANPALPSDFKTILD
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Telomerase is a ribonucleoprotein enzyme essential for the replication of chromosome termini in most eukaryotes. Active in progenitor and cancer cells. Inactive, or very low activity, in normal somatic cells. Catalytic component of the teleromerase holoenzyme complex whose main activity is the elongation of telomeres by acting as a reverse transcriptase that adds simple sequence repeats to chromosome ends by copying a template sequence within the RNA component of the enzyme. Catalyzes the RNA-dependent extension of 3'-chromosomal termini with the 6-nucleotide telomeric repeat unit, 5'-TTAGGG-3'. The catalytic cycle involves primer binding, primer extension and release of product once the template boundary has been reached or nascent product translocation followed by further extension. More active on substrates containing 2 or 3 telomeric repeats. Telomerase activity is regulated by a number of factors including telomerase complex-associated proteins, chaperones and polypeptide modifiers. Modulates Wnt signaling. Plays important roles in aging and antiapoptosis.
Phosphorylation at Tyr-707 under oxidative stress leads to translocation of TERT to the cytoplasm and reduces its antiapoptotic activity. Dephosphorylated by SHP2/PTPN11 leading to nuclear retention. Phosphorylation at Ser-227 by the AKT pathway promotes nuclear location. Phosphorylation at the G2/M phase at Ser-457 by DYRK2 promotes ubiquitination by the EDVP complex and degradation.
Ubiquitinated by the EDVP complex, a E3 ligase complex following phosphorylation at Ser-457 by DYRK2. Ubiquitinated leads to proteasomal degradation.
(Microbial infection) In case of infection by HIV-1, the EDVP complex is hijacked by HIV-1 via interaction between HIV-1 Vpr and DCAF1/VPRBP, leading to ubiquitination and degradation.
Nucleus>Nucleolus. Nucleus>Nucleoplasm. Nucleus. Chromosome>Telomere. Cytoplasm. Nucleus>PML body.
Note: Shuttling between nuclear and cytoplasm depends on cell cycle, phosphorylation states, transformation and DNA damage. Diffuse localization in the nucleoplasm. Enriched in nucleoli of certain cell types. Translocated to the cytoplasm via nuclear pores in a CRM1/RAN-dependent manner involving oxidative stress-mediated phosphorylation at Tyr-707. Dephosphorylation at this site by SHP2 retains TERT in the nucleus. Translocated to the nucleus by phosphorylation by AKT.
Expressed at a high level in thymocyte subpopulations, at an intermediate level in tonsil T-lymphocytes, and at a low to undetectable level in peripheral blood T-lymphocytes.
The primer grip sequence in the RT domain is required for telomerase activity and for stable association with short telomeric primers.
The RNA-interacting domain 1 (RD1)/N-terminal extension (NTE) is required for interaction with the pseudoknot-template domain of each of TERC dimers. It contains anchor sites that bind primer nucleotides upstream of the RNA-DNA hybrid and is thus an essential determinant of repeat addition processivity.
The RNA-interacting domain 2 (RD2) is essential for both interaction with the CR4-CR5 domain of TERC and for DNA synthesis.
Belongs to the reverse transcriptase family. Telomerase subfamily.
Research Fields
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Hepatocellular carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Gastric cancer. (View pathway)
References
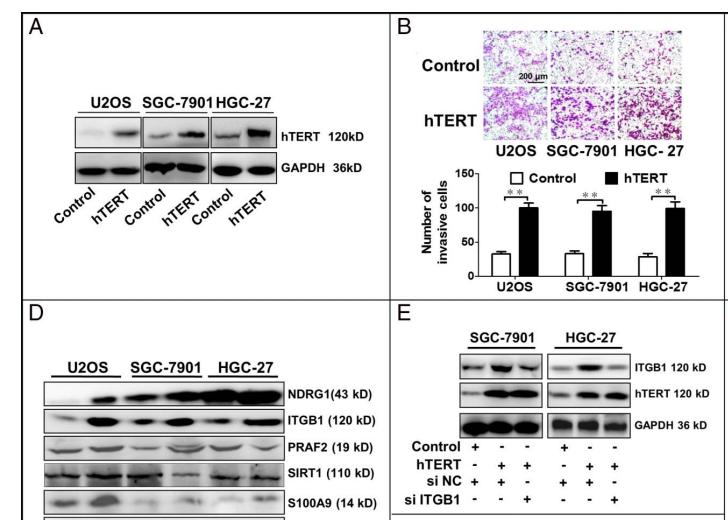
Application: WB Species: human Sample: human gastric cancer
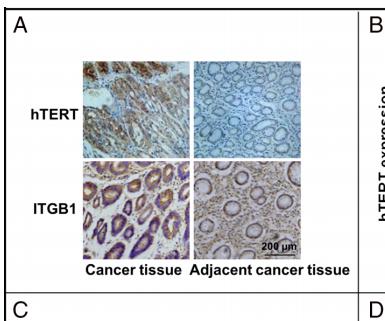
Application: IHC Species: human Sample: human gastric cancer
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.