PGC1 Antibody - #AF5395
| Product: | PGC1 Antibody |
| Catalog: | AF5395 |
| Description: | Rabbit polyclonal antibody to PGC1 |
| Application: | WB IHC |
| Cited expt.: | WB, IHC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 90~140 kD; 91kD(Calculated). |
| Uniprot: | Q9UBK2 |
| RRID: | AB_2837880 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF5395, RRID:AB_2837880.
Fold/Unfold
L PGC 1alpha;LEM6;Ligand effect modulator 6;Peroxisome proliferative activated receptor gamma coactivator 1 alpha;Peroxisome proliferative activated receptor gamma coactivator 1;Peroxisome proliferator activated receptor gamma coactivator 1 alpha;Peroxisome proliferator activated receptor gamma coactivator 1 alpha transcript variant B4 3ext;Peroxisome proliferator activated receptor gamma coactivator 1 alpha transcript variant B4 8a;Peroxisome proliferator activated receptor gamma coactivator 1 alpha transcript variant B4;Peroxisome proliferator activated receptor gamma coactivator 1 alpha transcript variant B5;Peroxisome proliferator activated receptor gamma coactivator 1 alpha transcript variant B5 NT;Peroxisome proliferator-activated receptor gamma coactivator 1-alpha;PGC 1 (alpha);PGC 1 alpha;PGC 1v;PGC-1-alpha;PGC1;PGC1(alpha);PGC1A;PGC1v;PPAR gamma coactivator 1 alpha 3 ligand effect modulator 6;PPAR gamma coactivator 1 alpha;PPAR gamma coactivator 1;PPAR gamma coactivator variant form;PPAR-gamma coactivator 1-alpha;PPARGC 1 alpha;PPARGC-1-alpha;PPARGC1;PPARGC1A;PRGC1_HUMAN;
Immunogens
A synthesized peptide derived from human PGC1 alpha, corresponding to a region within the internal amino acids.
Heart, skeletal muscle, liver and kidney. Expressed at lower levels in brain and pancreas and at very low levels in the intestine and white adipose tissue. In skeletal muscle, levels were lower in obese than in lean subjects and fasting induced a 2-fold increase in levels in the skeletal muscle in obese subjects.
- Q9UBK2 PRGC1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAWDMCNQDSESVWSDIECAALVGEDQPLCPDLPELDLSELDVNDLDTDSFLGGLKWCSDQSEIISNQYNNEPSNIFEKIDEENEANLLAVLTETLDSLPVDEDGLPSFDALTDGDVTTDNEASPSSMPDGTPPPQEAEEPSLLKKLLLAPANTQLSYNECSGLSTQNHANHNHRIRTNPAIVKTENSWSNKAKSICQQQKPQRRPCSELLKYLTTNDDPPHTKPTENRNSSRDKCTSKKKSHTQSQSQHLQAKPTTLSLPLTPESPNDPKGSPFENKTIERTLSVELSGTAGLTPPTTPPHKANQDNPFRASPKLKSSCKTVVPPPSKKPRYSESSGTQGNNSTKKGPEQSELYAQLSKSSVLTGGHEERKTKRPSLRLFGDHDYCQSINSKTEILINISQELQDSRQLENKDVSSDWQGQICSSTDSDQCYLRETLEASKQVSPCSTRKQLQDQEIRAELNKHFGHPSQAVFDDEADKTGELRDSDFSNEQFSKLPMFINSGLAMDGLFDDSEDESDKLSYPWDGTQSYSLFNVSPSCSSFNSPCRDSVSPPKSLFSQRPQRMRSRSRSFSRHRSCSRSPYSRSRSRSPGSRSSSRSCYYYESSHYRHRTHRNSPLYVRSRSRSPYSRRPRYDSYEEYQHERLKREEYRREYEKRESERAKQRERQRQKAIEERRVIYVGKIRPDTTRTELRDRFEVFGEIEECTVNLRDDGDSYGFITYRYTCDAFAALENGYTLRRSNETDFELYFCGRKQFFKSNYADLDSNSDDFDPASTKSKYDSLDFDSLLKEAQRSLRR
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Transcriptional coactivator for steroid receptors and nuclear receptors. Greatly increases the transcriptional activity of PPARG and thyroid hormone receptor on the uncoupling protein promoter. Can regulate key mitochondrial genes that contribute to the program of adaptive thermogenesis. Plays an essential role in metabolic reprogramming in response to dietary availability through coordination of the expression of a wide array of genes involved in glucose and fatty acid metabolism. Induces the expression of PERM1 in the skeletal muscle in an ESRRA-dependent manner. Also involved in the integration of the circadian rhythms and energy metabolism. Required for oscillatory expression of clock genes, such as ARNTL/BMAL1 and NR1D1, through the coactivation of RORA and RORC, and metabolic genes, such as PDK4 and PEPCK.
Phosphorylation by AMPK in skeletal muscle increases activation of its own promoter. Phosphorylated by CLK2.
Heavily acetylated by GCN5 and biologically inactive under conditions of high nutrients. Deacetylated by SIRT1 in low nutrients/high NAD conditions.
Ubiquitinated. Ubiquitination by RNF34 induces proteasomal degradation.
Nucleus. Nucleus>PML body.
Nucleus.
Cytoplasm. Nucleus.
Nucleus. Nucleus>PML body.
Nucleus.
Heart, skeletal muscle, liver and kidney. Expressed at lower levels in brain and pancreas and at very low levels in the intestine and white adipose tissue. In skeletal muscle, levels were lower in obese than in lean subjects and fasting induced a 2-fold increase in levels in the skeletal muscle in obese subjects.
Research Fields
· Environmental Information Processing > Signal transduction > AMPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Apelin signaling pathway. (View pathway)
· Human Diseases > Endocrine and metabolic diseases > Insulin resistance.
· Human Diseases > Neurodegenerative diseases > Huntington's disease.
· Organismal Systems > Aging > Longevity regulating pathway. (View pathway)
· Organismal Systems > Endocrine system > Insulin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Adipocytokine signaling pathway.
· Organismal Systems > Endocrine system > Glucagon signaling pathway.
References
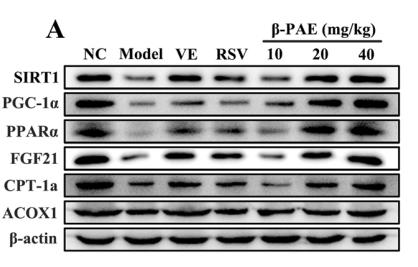
Application: WB Species: Rat Sample: livers
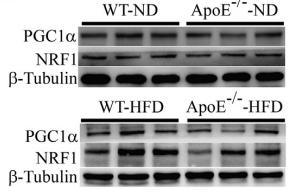
Application: WB Species: Mouse Sample: endothelial cells
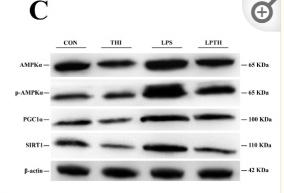
Application: WB Species: Goat Sample: RECs
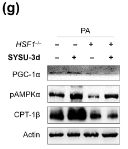
Application: WB Species: Human Sample: L02 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.