N Cadherin Antibody - #AF5239
| Product: | N Cadherin Antibody |
| Catalog: | AF5239 |
| Description: | Rabbit polyclonal antibody to N Cadherin |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IHC, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 100~150kd; 100kD(Calculated). |
| Uniprot: | P19022 |
| RRID: | AB_2837725 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF5239, RRID:AB_2837725.
Fold/Unfold
CADH2_HUMAN; Cadherin 2; Cadherin 2 N cadherin neuronal; Cadherin 2 type 1; Cadherin 2 type 1 N cadherin neuronal; Cadherin 2, type 1, N-cadherin (neuronal); Cadherin-2; Cadherin2; Calcium dependent adhesion protein neuronal; CD325; CD325 antigen; CDH2; CDHN; CDw325; CDw325 antigen; N cadherin 1; N-cadherin; NCAD; Neural cadherin; OTTHUMP00000066304; OTTHUMP00000067378;
Immunogens
A synthesized peptide derived from human N Cadherin, corresponding to a region within C-terminal amino acids.
- P19022 CADH2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MCRIAGALRTLLPLLAALLQASVEASGEIALCKTGFPEDVYSAVLSKDVHEGQPLLNVKFSNCNGKRKVQYESSEPADFKVDEDGMVYAVRSFPLSSEHAKFLIYAQDKETQEKWQVAVKLSLKPTLTEESVKESAEVEEIVFPRQFSKHSGHLQRQKRDWVIPPINLPENSRGPFPQELVRIRSDRDKNLSLRYSVTGPGADQPPTGIFIINPISGQLSVTKPLDREQIARFHLRAHAVDINGNQVENPIDIVINVIDMNDNRPEFLHQVWNGTVPEGSKPGTYVMTVTAIDADDPNALNGMLRYRIVSQAPSTPSPNMFTINNETGDIITVAAGLDREKVQQYTLIIQATDMEGNPTYGLSNTATAVITVTDVNDNPPEFTAMTFYGEVPENRVDIIVANLTVTDKDQPHTPAWNAVYRISGGDPTGRFAIQTDPNSNDGLVTVVKPIDFETNRMFVLTVAAENQVPLAKGIQHPPQSTATVSVTVIDVNENPYFAPNPKIIRQEEGLHAGTMLTTFTAQDPDRYMQQNIRYTKLSDPANWLKIDPVNGQITTIAVLDRESPNVKNNIYNATFLASDNGIPPMSGTGTLQIYLLDINDNAPQVLPQEAETCETPDPNSINITALDYDIDPNAGPFAFDLPLSPVTIKRNWTITRLNGDFAQLNLKIKFLEAGIYEVPIIITDSGNPPKSNISILRVKVCQCDSNGDCTDVDRIVGAGLGTGAIIAILLCIIILLILVLMFVVWMKRRDKERQAKQLLIDPEDDVRDNILKYDEEGGGEEDQDYDLSQLQQPDTVEPDAIKPVGIRRMDERPIHAEPQYPVRSAAPHPGDIGDFINEGLKAADNDPTAPPYDSLLVFDYEGSGSTAGSLSSLNSSSSGGEQDYDYLNDWGPRFKKLADMYGGGDD
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Calcium-dependent cell adhesion protein; preferentially mediates homotypic cell-cell adhesion by dimerization with a CDH2 chain from another cell. Cadherins may thus contribute to the sorting of heterogeneous cell types. Acts as a regulator of neural stem cells quiescence by mediating anchorage of neural stem cells to ependymocytes in the adult subependymal zone: upon cleavage by MMP24, CDH2-mediated anchorage is affected, leading to modulate neural stem cell quiescence. CDH2 may be involved in neuronal recognition mechanism. In hippocampal neurons, may regulate dendritic spine density.
Cleaved by MMP24. Ectodomain cleavage leads to the generation of a soluble 90 kDa amino-terminal soluble fragment and a 45 kDa membrane-bound carboxy-terminal fragment 1 (CTF1), which is further cleaved by gamma-secretase into a 35 kDa. Cleavage in neural stem cells by MMP24 affects CDH2-mediated anchorage of neural stem cells to ependymocytes in the adult subependymal zone, leading to modulate neural stem cell quiescence (By similarity).
May be phosphorylated by OBSCN.
Cell membrane>Single-pass type I membrane protein. Cell membrane>Sarcolemma. Cell junction. Cell surface.
Note: Colocalizes with TMEM65 at the intercalated disk in cardiomyocytes. Colocalizes with OBSCN at the intercalated disk and at sarcolemma in cardiomyocytes.
Three calcium ions are usually bound at the interface of each cadherin domain and rigidify the connections, imparting a strong curvature to the full-length ectodomain. Calcium-binding sites are occupied sequentially in the order of site 3, then site 2 and site 1.
Research Fields
· Environmental Information Processing > Signaling molecules and interaction > Cell adhesion molecules (CAMs). (View pathway)
· Human Diseases > Cardiovascular diseases > Arrhythmogenic right ventricular cardiomyopathy (ARVC).
References
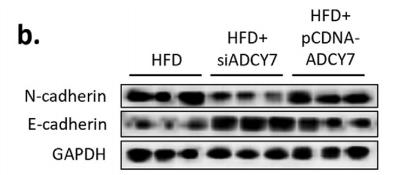
Application: WB Species: Human Sample: 5-8 F and HNE1 cells
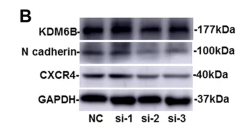
Application: WB Species: Human Sample: BMSCs
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.