SIRT3 Antibody - #AF5135
| Product: | SIRT3 Antibody |
| Catalog: | AF5135 |
| Description: | Rabbit polyclonal antibody to SIRT3 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IHC, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Sheep, Rabbit |
| Mol.Wt.: | 29 kDa; 44kD(Calculated). |
| Uniprot: | Q9NTG7 |
| RRID: | AB_2837621 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF5135, RRID:AB_2837621.
Fold/Unfold
hSIRT 3; hSIRT3; Mitochondrial nicotinamide adenine dinucleotide dependent deacetylase; NAD dependent deacetylase sirtuin 3 mitochondrial; NAD-dependent protein deacetylase sirtuin-3, mitochondrial; Regulatory protein SIR2 homolog 3; Silent mating type information regulation 2 S.cerevisiae homolog 3; Sir 2 like 3; SIR 2 like protein 3; SIR 3; SIR2 L3; Sir2 like 3; SIR2 like protein 3; SIR2-like protein 3; SIR2L3; SIR3_HUMAN; SIRT 3; SIRT3; Sirtuin 3; Sirtuin silent mating type information regulation 2 homolog 3 (S. cerevisiae); Sirtuin type 3; Sirtuin3;
Immunogens
A synthesized peptide derived from human SIRT3, corresponding to a region within C-terminal amino acids.
- Q9NTG7 SIR3_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAFWGWRAAAALRLWGRVVERVEAGGGVGPFQACGCRLVLGGRDDVSAGLRGSHGARGEPLDPARPLQRPPRPEVPRAFRRQPRAAAPSFFFSSIKGGRRSISFSVGASSVVGSGGSSDKGKLSLQDVAELIRARACQRVVVMVGAGISTPSGIPDFRSPGSGLYSNLQQYDLPYPEAIFELPFFFHNPKPFFTLAKELYPGNYKPNVTHYFLRLLHDKGLLLRLYTQNIDGLERVSGIPASKLVEAHGTFASATCTVCQRPFPGEDIRADVMADRVPRCPVCTGVVKPDIVFFGEPLPQRFLLHVVDFPMADLLLILGTSLEVEPFASLTEAVRSSVPRLLINRDLVGPLAWHPRSRDVAQLGDVVHGVESLVELLGWTEEMRDLVQRETGKLDGPDK
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
NAD-dependent protein deacetylase. Activates or deactivates mitochondrial target proteins by deacetylating key lysine residues. Known targets include ACSS1, IDH, GDH, SOD2, PDHA1, LCAD, SDHA and the ATP synthase subunit ATP5PO. Contributes to the regulation of the cellular energy metabolism. Important for regulating tissue-specific ATP levels. In response to metabolic stress, deacetylates transcription factor FOXO3 and recruits FOXO3 and mitochondrial RNA polymerase POLRMT to mtDNA to promote mtDNA transcription. Acts as a regulator of ceramide metabolism by mediating deacetylation of ceramide synthases CERS1, CERS2 and CERS6, thereby increasing their activity and promoting mitochondrial ceramide accumulation (By similarity).
Processed by mitochondrial processing peptidase (MPP) to give a 28 kDa product. Such processing is probably essential for its enzymatic activity.
Mitochondrion matrix.
Widely expressed.
Belongs to the sirtuin family. Class I subfamily.
Research Fields
· Human Diseases > Cancers: Overview > Central carbon metabolism in cancer. (View pathway)
References
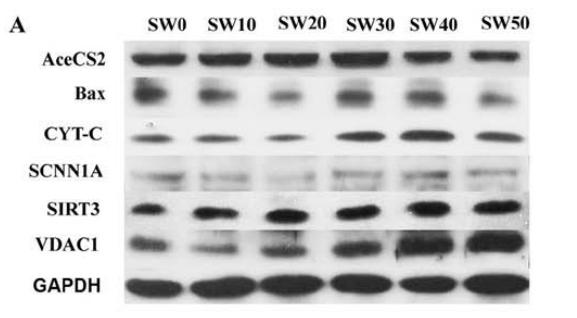
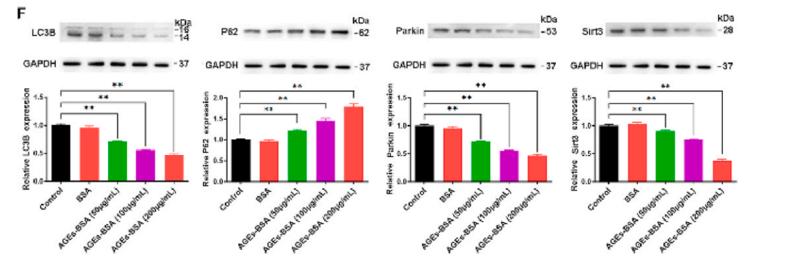
Application: WB Species: mice Sample: bone marrow mesenchymal stem (BMSCs)
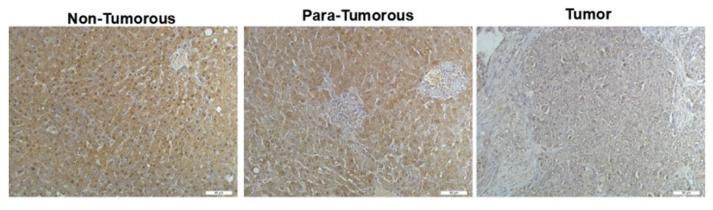
Application: IF/ICC Species: Mouse Sample:
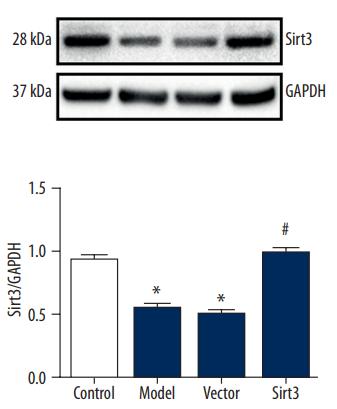
Application: WB Species: Rat Sample: H9C2 cell
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.