CD63 Antibody - #AF5117
| Product: | CD63 Antibody |
| Catalog: | AF5117 |
| Description: | Rabbit polyclonal antibody to CD63 |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Mol.Wt.: | 25kDa,47kDa; 26kD(Calculated). |
| Uniprot: | P08962 |
| RRID: | AB_2837603 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF5117, RRID:AB_2837603.
Fold/Unfold
Lysosomal associated membrane protein 3; CD 63; CD63; CD63 antigen (melanoma 1 antigen); CD63 antigen; CD63 antigen melanoma 1 antigen; CD63 molecule; CD63_HUMAN; gp55; Granulophysin; LAMP 3; LAMP-3; LAMP3; LIMP; Lysosomal-associated membrane protein 3; Lysosome associated membrane glycoprotein 3; Mast cell antigen AD1; ME491; Melanoma 1 antigen; Melanoma associated antigen ME491; Melanoma associated antigen MLA1; Melanoma-associated antigen ME491; MGC72893; MLA 1; MLA1; NGA; Ocular melanoma associated antigen; Ocular melanoma-associated antigen; OMA81H; PTLGP40; Tetraspanin 30; Tetraspanin-30; Tspan 30; Tspan-30; TSPAN30;
Immunogens
A synthesized peptide derived from human CD63
Detected in platelets (at protein level). Dysplastic nevi, radial growth phase primary melanomas, hematopoietic cells, tissue macrophages.
- P08962 CD63_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAVEGGMKCVKFLLYVLLLAFCACAVGLIAVGVGAQLVLSQTIIQGATPGSLLPVVIIAVGVFLFLVAFVGCCGACKENYCLMITFAIFLSLIMLVEVAAAIAGYVFRDKVMSEFNNNFRQQMENYPKNNHTASILDRMQADFKCCGAANYTDWEKIPSMSKNRVPDSCCINVTVGCGINFNEKAIHKEGCVEKIGGWLRKNVLVVAAAALGIAFVEVLGIVFACCLVKSIRSGYEVM
Research Backgrounds
Functions as cell surface receptor for TIMP1 and plays a role in the activation of cellular signaling cascades. Plays a role in the activation of ITGB1 and integrin signaling, leading to the activation of AKT, FAK/PTK2 and MAP kinases. Promotes cell survival, reorganization of the actin cytoskeleton, cell adhesion, spreading and migration, via its role in the activation of AKT and FAK/PTK2. Plays a role in VEGFA signaling via its role in regulating the internalization of KDR/VEGFR2. Plays a role in intracellular vesicular transport processes, and is required for normal trafficking of the PMEL luminal domain that is essential for the development and maturation of melanocytes. Plays a role in the adhesion of leukocytes onto endothelial cells via its role in the regulation of SELP trafficking. May play a role in mast cell degranulation in response to Ms4a2/FceRI stimulation, but not in mast cell degranulation in response to other stimuli.
Palmitoylated at a low, basal level in unstimulated platelets. The level of palmitoylation increases when platelets are activated by thrombin (in vitro).
Cell membrane>Multi-pass membrane protein. Lysosome membrane>Multi-pass membrane protein. Late endosome membrane>Multi-pass membrane protein. Endosome>Multivesicular body. Melanosome. Secreted>Extracellular exosome. Cell surface.
Note: Also found in Weibel-Palade bodies of endothelial cells (PubMed:10793155). Located in platelet dense granules (PubMed:7682577). Detected in a subset of pre-melanosomes. Detected on intralumenal vesicles (ILVs) within multivesicular bodies (PubMed:21962903).
Detected in platelets (at protein level). Dysplastic nevi, radial growth phase primary melanomas, hematopoietic cells, tissue macrophages.
Belongs to the tetraspanin (TM4SF) family.
Research Fields
· Cellular Processes > Transport and catabolism > Lysosome. (View pathway)
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
References
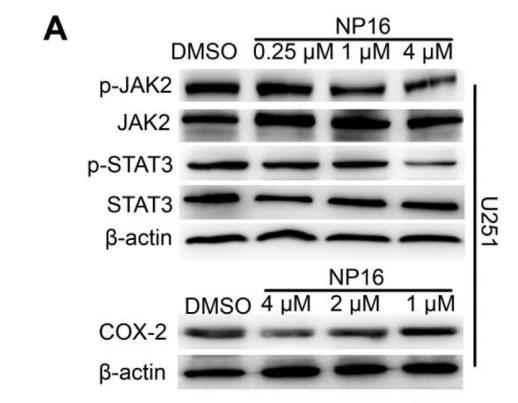
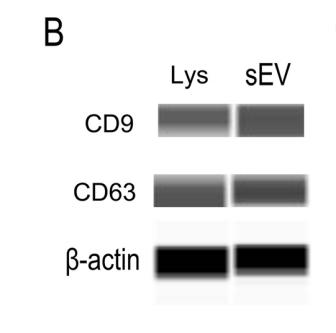
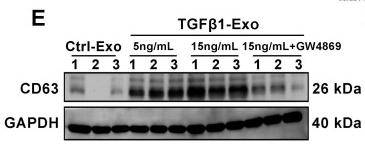
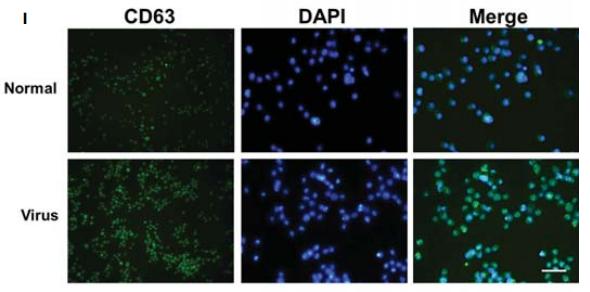
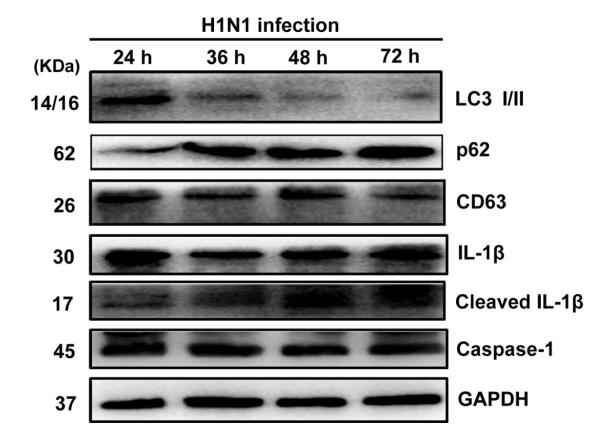
Application: WB Species: Mouse Sample:
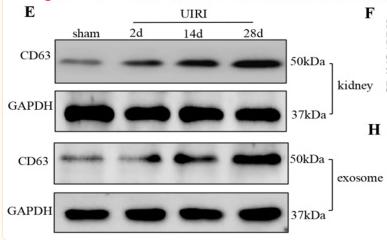
Application: WB Species: Mice Sample: NRK-52E cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.