XBP1 Antibody - #AF5110
| Product: | XBP1 Antibody |
| Catalog: | AF5110 |
| Description: | Rabbit polyclonal antibody to XBP1 |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Sheep, Dog |
| Mol.Wt.: | 30~45kD(Unspliced),55kD(Spliced).; 29kD(Calculated). |
| Uniprot: | P17861 |
| RRID: | AB_2837596 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF5110, RRID:AB_2837596.
Fold/Unfold
Tax responsive element binding protein 5; Tax-responsive element-binding protein 5; TREB5; X box binding protein 1; X box binding protein 2; X-box-binding protein 1; XBP 1; XBP-1; XBP1; XBP1_HUMAN; XBP2;
Immunogens
A synthesized peptide derived from human XBP1, corresponding to a region within C-terminal amino acids.
Expressed in plasma cells in rheumatoid synovium (PubMed:11460154). Over-expressed in primary breast cancer and metastatic breast cancer cells (PubMed:25280941). Isoform 1 and isoform 2 are expressed at higher level in proliferating as compared to confluent quiescent endothelial cells (PubMed:19416856).
- P17861 XBP1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MVVVAAAPNPADGTPKVLLLSGQPASAAGAPAGQALPLMVPAQRGASPEAASGGLPQARKRQRLTHLSPEEKALRRKLKNRVAAQTARDRKKARMSELEQQVVDLEEENQKLLLENQLLREKTHGLVVENQELRQRLGMDALVAEEEAEAKGNEVRPVAGSAESAALRLRAPLQQVQAQLSPLQNISPWILAVLTLQIQSLISCWAFWTTWTQSCSSNALPQSLPAWRSSQRSTQKDPVPYQPPFLCQWGRHQPSWKPLMN
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Functions as a transcription factor during endoplasmic reticulum (ER) stress by regulating the unfolded protein response (UPR). Required for cardiac myogenesis and hepatogenesis during embryonic development, and the development of secretory tissues such as exocrine pancreas and salivary gland (By similarity). Involved in terminal differentiation of B lymphocytes to plasma cells and production of immunoglobulins. Modulates the cellular response to ER stress in a PIK3R-dependent manner. Binds to the cis-acting X box present in the promoter regions of major histocompatibility complex class II genes. Involved in VEGF-induced endothelial cell (EC) proliferation and retinal blood vessel formation during embryonic development but also for angiogenesis in adult tissues under ischemic conditions. Functions also as a major regulator of the UPR in obesity-induced insulin resistance and type 2 diabetes for the management of obesity and diabetes prevention (By similarity).
Plays a role in the unconventional cytoplasmic splicing processing of its own mRNA triggered by the endoplasmic reticulum (ER) transmembrane endoribonuclease ENR1: upon ER stress, the emerging XBP1 polypeptide chain, as part of a mRNA-ribosome-nascent chain (R-RNC) complex, cotranslationally recruits its own unprocessed mRNA through transient docking to the ER membrane and translational pausing, therefore facilitating efficient IRE1-mediated XBP1 mRNA isoform 2 production. In endothelial cells (EC), associated with KDR, promotes IRE1-mediated XBP1 mRNA isoform 2 productions in a vascular endothelial growth factor (VEGF)-dependent manner, leading to EC proliferation and angiogenesis. Functions as a negative feed-back regulator of the potent transcription factor XBP1 isoform 2 protein levels through proteasome-mediated degradation, thus preventing the constitutive activation of the ER stress response signaling pathway. Inhibits the transactivation activity of XBP1 isoform 2 in myeloma cells (By similarity). Acts as a weak transcriptional factor. Together with HDAC3, contributes to the activation of NFE2L2-mediated HMOX1 transcription factor gene expression in a PI(3)K/mTORC2/Akt-dependent signaling pathway leading to EC survival under disturbed flow/oxidative stress. Binds to the ER stress response element (ERSE) upon ER stress. Binds to the consensus 5'-GATGACGTG[TG]N(3)[AT]T-3' sequence related to cAMP responsive element (CRE)-like sequences. Binds the Tax-responsive element (TRE) present in the long terminal repeat (LTR) of T-cell leukemia virus type 1 (HTLV-I) and to the TPA response elements (TRE). Associates preferentially to the HDAC3 gene promoter region in a static flow-dependent manner. Binds to the CDH5/VE-cadherin gene promoter region.
Functions as a stress-inducible potent transcriptional activator during endoplasmic reticulum (ER) stress by inducing unfolded protein response (UPR) target genes via binding to the UPR element (UPRE). Up-regulates target genes encoding ER chaperones and ER-associated degradation (ERAD) components to enhance the capacity of productive folding and degradation mechanism, respectively, in order to maintain the homeostasis of the ER under ER stress. Plays a role in the production of immunoglobulins and interleukin-6 in the presence of stimuli required for plasma cell differentiation (By similarity). Induces phospholipid biosynthesis and ER expansion. Contributes to the VEGF-induced endothelial cell (EC) growth and proliferation in a Akt/GSK-dependent and/or -independent signaling pathway, respectively, leading to beta-catenin nuclear translocation and E2F2 gene expression. Promotes umbilical vein EC apoptosis and atherosclerotisis development in a caspase-dependent signaling pathway, and contributes to VEGF-induced EC proliferation and angiogenesis in adult tissues under ischemic conditions. Involved in the regulation of endostatin-induced autophagy in EC through BECN1 transcriptional activation. Plays a role as an oncogene by promoting tumor progression: stimulates zinc finger protein SNAI1 transcription to induce epithelial-to-mesenchymal (EMT) transition, cell migration and invasion of breast cancer cells. Involved in adipocyte differentiation by regulating lipogenic gene expression during lactation. Plays a role in the survival of both dopaminergic neurons of the substantia nigra pars compacta (SNpc), by maintaining protein homeostasis and of myeloma cells. Increases insulin sensitivity in the liver as a response to a high carbohydrate diet, resulting in improved glucose tolerance. Improves also glucose homeostasis in an ER stress- and/or insulin-independent manner through both binding and proteasome-induced degradation of the transcription factor FOXO1, hence resulting in suppression of gluconeogenic genes expression and in a reduction of blood glucose levels. Controls the induction of de novo fatty acid synthesis in hepatocytes by regulating the expression of a subset of lipogenic genes in an ER stress- and UPR-independent manner (By similarity). Associates preferentially to the HDAC3 gene promoter region in a disturbed flow-dependent manner. Binds to the BECN1 gene promoter region. Binds to the CDH5/VE-cadherin gene promoter region. Binds to the ER stress response element (ERSE) upon ER stress. Binds to the 5'-CCACG-3' motif in the PPARG promoter (By similarity).
Isoform 2 is acetylated by EP300; acetylation positively regulates the transcriptional activity of XBP1 isoform 2. Isoform 2 is deacetylated by SIRT1; deacetylation negatively regulates the transcriptional activity of XBP1 isoform 2.
Isoform 1 is ubiquitinated, leading to proteasome-mediated degradation in response to ER stress.
X-box-binding protein 1, cytoplasmic form and luminal form are produced by intramembrane proteolytic cleavage of ER membrane-anchored isoform 1 triggered by HM13/SPP in a DERL1-RNF139-dependent and VCP/p97-independent manner. X-box-binding protein 1, luminal form is ubiquitinated leading to proteasomal degradation.
Endoplasmic reticulum.
Note: Colocalizes with ERN1 and KDR in the endoplasmic reticulum in endothelial cells in a vascular endothelial growth factor (VEGF)-dependent manner (PubMed:23529610).
Nucleus. Cytoplasm. Endoplasmic reticulum membrane>Single-pass type II membrane protein. Endoplasmic reticulum membrane>Peripheral membrane protein. Membrane>Peripheral membrane protein.
Note: Shows no preferential localization to either the nucleus or the cytoplasm (By similarity). Shuttles between the nucleus and the cytoplasm in a CRM1-dependent manner (PubMed:16461360). Localizes predominantly at the endoplasmic reticulum membrane as a membrane-spanning protein; whereas may be only marginally localized on the cytosolic side of the ER membrane as a peripheral membrane (PubMed:19394296, PubMed:25190803).
Nucleus. Cytoplasm.
Note: Localizes predominantly in the nucleus. Colocalizes in the nucleus with SIRT1. Translocates into the nucleus in a PIK3R-, ER stress-induced- and/or insulin-dependent manner (By similarity).
Cytoplasm. Nucleus.
Note: Localizes in the cytoplasm and nucleus after HM13/SPP-mediated intramembranaire proteolytic cleavage of isoform 1 (PubMed:25239945).
Expressed in plasma cells in rheumatoid synovium. Over-expressed in primary breast cancer and metastatic breast cancer cells. Isoform 1 and isoform 2 are expressed at higher level in proliferating as compared to confluent quiescent endothelial cells.
Isoform 1 and isoform 2 N-terminus domains are necessary for nuclear localization targeting. Isoform 1 C-terminus domain confers localization to the cytoplasm and is sufficient to impose rapid degradation (By similarity). Isoform 1 transmembrane signal-anchor domain is necessary for its own mRNA to be recruited to the endoplasmic reticulum (ER) which will undergo unconventional ERN1-dependent splicing in response to ER stress (PubMed:19394296, PubMed:21233347). Isoform 1 N-terminus and C-terminus regions are necessary for DNA-binding and weak transcriptional activity, respectively. Isoform 2 N-terminus and C-terminus regions are necessary for DNA-binding and strong transcriptional activity upon ER stress, respectively (PubMed:11779464, PubMed:8657566). Isoform 2 C-terminus region contains a nuclear exclusion signal (NES) at positions 186 through 208. Isoform 2 C-terminus region contains a degradation domain at positions 209 through 261 (PubMed:16461360).
Belongs to the bZIP family.
Research Fields
· Genetic Information Processing > Folding, sorting and degradation > Protein processing in endoplasmic reticulum. (View pathway)
· Human Diseases > Endocrine and metabolic diseases > Non-alcoholic fatty liver disease (NAFLD).
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
References
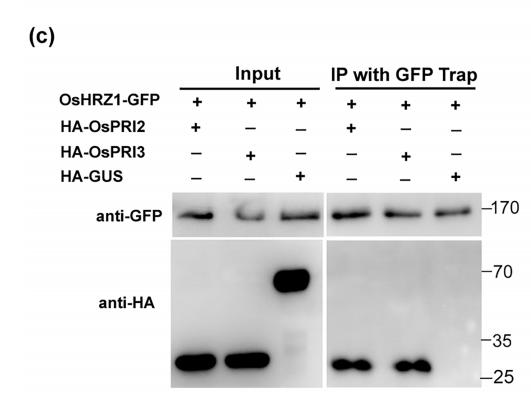
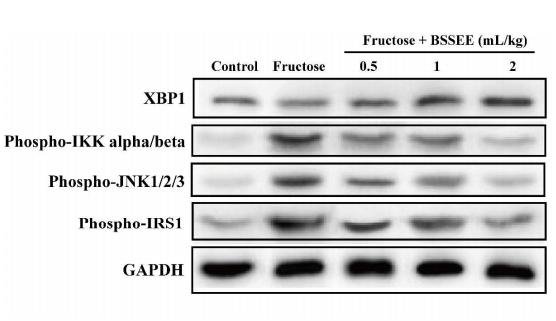
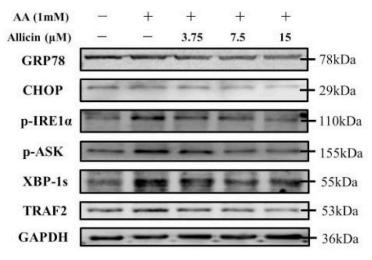
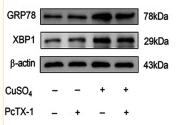
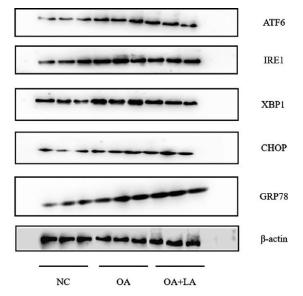
Application: WB Species: human Sample: Molm13 cell
Application: WB Species: mouse Sample: Liver
Application: WB Species: rat Sample:
Application: WB Species: Rat Sample: HSC-T6 cells
Application: WB Species: Human Sample: HepG2 cells
Application: WB Species: Mouse Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.