NEK7 Antibody - #DF4467
| Product: | NEK7 Antibody |
| Catalog: | DF4467 |
| Description: | Rabbit polyclonal antibody to NEK7 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IHC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 35 KD; 35kD(Calculated). |
| Uniprot: | Q8TDX7 |
| RRID: | AB_2836822 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF4467, RRID:AB_2836822.
Fold/Unfold
EC 2.7.11.1; NEK7; NEK7_HUMAN; Never in mitosis A-related kinase 7; Never in mitosis gene a-related kinase 7; NIMA (never in mitosis gene a) related kinase 7; NIMA (never in mitosis gene a)-related expressed kinase 7; NimA related protein kinase 7; NIMA-related kinase 7; NimA-related protein kinase 7; Serine/threonine-protein kinase Nek7;
Immunogens
A synthesized peptide derived from human NEK7, corresponding to a region within the internal amino acids.
Highly expressed in lung, muscle, testis, brain, heart, liver, leukocyte and spleen. Lower expression in ovary, prostate and kidney. No expression seen in small intestine.
- Q8TDX7 NEK7_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MDEQSQGMQGPPVPQFQPQKALRPDMGYNTLANFRIEKKIGRGQFSEVYRAACLLDGVPVALKKVQIFDLMDAKARADCIKEIDLLKQLNHPNVIKYYASFIEDNELNIVLELADAGDLSRMIKHFKKQKRLIPERTVWKYFVQLCSALEHMHSRRVMHRDIKPANVFITATGVVKLGDLGLGRFFSSKTTAAHSLVGTPYYMSPERIHENGYNFKSDIWSLGCLLYEMAALQSPFYGDKMNLYSLCKKIEQCDYPPLPSDHYSEELRQLVNMCINPDPEKRPDVTYVYDVAKRMHACTASS
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Protein kinase which plays an important role in mitotic cell cycle progression. Required for microtubule nucleation activity of the centrosome, robust mitotic spindle formation and cytokinesis. Phosphorylates RPS6KB1.
Phosphorylation at Ser-195 required for its activation.
Nucleus. Cytoplasm. Cytoplasm>Cytoskeleton>Spindle pole. Cytoplasm>Cytoskeleton>Microtubule organizing center>Centrosome.
Note: Present at centrosome throughout the cell cycle (PubMed:17586473). Also detected at spindle midzone of the anaphase cells and eventually concentrates at the midbody (PubMed:17586473). Interaction with ANKS3 prevents its translocation to the nucleus (By similarity).
Highly expressed in lung, muscle, testis, brain, heart, liver, leukocyte and spleen. Lower expression in ovary, prostate and kidney. No expression seen in small intestine.
Displays an autoinhibited conformation: Tyr-97 side chain points into the active site, interacts with the activation loop, and blocks the alphaC helix. The autoinhibitory conformation is released upon binding with NEK9.
The NTE (N-terminal extension) motif is a structural component of the catalytic domain and thus contributes to activity.
Belongs to the protein kinase superfamily. NEK Ser/Thr protein kinase family. NIMA subfamily.
Research Fields
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
References
Application: WB Species: Human Sample: colon cancer tissues
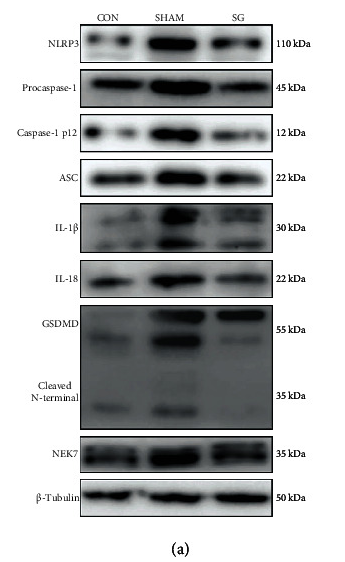
Application: WB Species: Rat Sample: diabetic myocardium
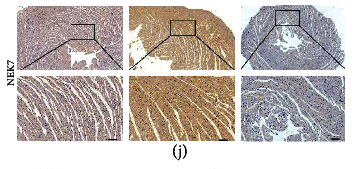
Application: IHC Species: Rat Sample: diabetic myocardium
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.