BOK Antibody - #DF3829
| Product: | BOK Antibody |
| Catalog: | DF3829 |
| Description: | Rabbit polyclonal antibody to BOK |
| Application: | WB IHC IF/ICC |
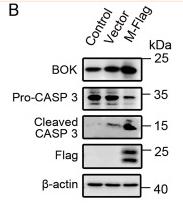
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Chicken, Xenopus |
| Mol.Wt.: | 23 KD; 23kD(Calculated). |
| Uniprot: | Q9UMX3 |
| RRID: | AB_2836186 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF3829, RRID:AB_2836186.
Fold/Unfold
Bcl 2 related ovarian killer protein; Bcl-2-like protein 9; Bcl-2-related ovarian killer protein; BCL2 related ovarian killer; Bcl2-L-9; BCL2L9; BOK; BOK_HUMAN; BOKL; Hbok; MGC4631;
Immunogens
A synthesized peptide derived from human BOK, corresponding to a region within N-terminal amino acids.
Expressed mainly in oocytes; weak expression in granulosa cells of the developing follicles. In adult human ovaries, expressed in granulosa cells at all follicular stages, but expression in primordial/primary follicles granulosa cell is stronger than in secondary and antral follicles.
- Q9UMX3 BOK_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MEVLRRSSVFAAEIMDAFDRSPTDKELVAQAKALGREYVHARLLRAGLSWSAPERAAPVPGRLAEVCAVLLRLGDELEMIRPSVYRNVARQLHISLQSEPVVTDAFLAVAGHIFSAGITWGKVVSLYAVAAGLAVDCVRQAQPAMVHALVDCLGEFVRKTLATWLRRRGGWTDVLKCVVSTDPGLRSHWLVAALCSFGRFLKAAFFVLLPER
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Apoptosis regulator that functions through different apoptotic signaling pathways. Plays a roles as pro-apoptotic protein that positively regulates intrinsic apoptotic process in a BAX- and BAK1-dependent manner or in a BAX- and BAK1-independent manner. In response to endoplasmic reticulum stress promotes mitochondrial apoptosis through downstream BAX/BAK1 activation and positive regulation of PERK-mediated unfolded protein response (By similarity). Activates apoptosis independently of heterodimerization with survival-promoting BCL2 and BCL2L1 through induction of mitochondrial outer membrane permeabilization, in a BAX- and BAK1-independent manner, in response to inhibition of ERAD-proteasome degradation system, resulting in cytochrome c release. In response to DNA damage, mediates intrinsic apoptotic process in a TP53-dependent manner. Plays a role in granulosa cell apoptosis by CASP3 activation. Plays a roles as anti-apoptotic protein during neuronal apoptotic process, by negatively regulating poly ADP-ribose polymerase-dependent cell death through regulation of neuronal calcium homeostasis and mitochondrial bioenergetics in response to NMDA excitation (By similarity). In addition to its role in apoptosis, may regulate trophoblast cell proliferation during the early stages of placental development, by acting on G1/S transition through regulation of CCNE1 expression. May also play a role as an inducer of autophagy by disrupting interaction between MCL1 and BECN1.
Pro-apoptotic molecule exerting its function through the mitochondrial pathway.
Ubiquitinated by AMFR/gp78 E3 ubiquitin ligase complex; mediates degradation by ubiquitin-proteasome pathway in a VCP/p97-dependent manner; prevents from pro-apoptotic activity; promotes degradation of newly synthesized proteins that are not ITPR1 associated.
Mitochondrion membrane>Single-pass membrane protein. Endoplasmic reticulum membrane>Single-pass membrane protein. Mitochondrion inner membrane. Cytoplasm. Nucleus. Mitochondrion. Endoplasmic reticulum. Mitochondrion outer membrane. Early endosome membrane. Recycling endosome membrane. Nucleus outer membrane. Golgi apparatus>cis-Golgi network membrane. Golgi apparatus>trans-Golgi network membrane. Membrane.
Note: Nuclear and cytoplasmic compartments in the early stages of apoptosis and during apoptosis it associates with mitochondria (PubMed:19942931). In healthy cells, associates loosely with the membrane in a hit-and-run mode. The insertion and accumulation on membranes is enhanced through the activity of death signals, resulting in the integration of the membrane-bound protein into the membrane (PubMed:15868100). The transmembrane domain controls subcellular localization; constitutes a tail-anchor. Localizes in early and late endosome upon blocking of apoptosis. Must localize to the mitochondria to induce mitochondrial outer membrane permeabilization and apoptosis (By similarity).
Membrane. Cytoplasm.
Expressed mainly in oocytes; weak expression in granulosa cells of the developing follicles. In adult human ovaries, expressed in granulosa cells at all follicular stages, but expression in primordial/primary follicles granulosa cell is stronger than in secondary and antral follicles.
BH4 domain mediates interaction with ITPR1.
Belongs to the Bcl-2 family.
Research Fields
· Cellular Processes > Cell growth and death > Apoptosis - multiple species. (View pathway)
References
Application: WB Species: Mouse Sample: lung tissue
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.