ATG16L1 Antibody - #DF3825
| Product: | ATG16L1 Antibody |
| Catalog: | DF3825 |
| Description: | Rabbit polyclonal antibody to ATG16L1 |
| Application: | WB IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog |
| Mol.Wt.: | 69 KD; 68kD(Calculated). |
| Uniprot: | Q676U5 |
| RRID: | AB_2836182 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF3825, RRID:AB_2836182.
Fold/Unfold
A16L1_HUMAN; APG16 like 1; APG16-like 1; APG16L; APG16L beta; ATG16 autophagy related 16 like 1; ATG16 autophagy related 16-like 1 (S. cerevisiae); ATG16A; ATG16L; Atg16l1; Autophagy related protein 16 1; Autophagy-related protein 16-1; FLJ00045; FLJ10035; FLJ10828; FLJ22677; IBD10; OTTHUMP00000164391; OTTHUMP00000164393; OTTHUMP00000165876; OTTHUMP00000165877; WD repeat domain 30; WDR30;
Immunogens
A synthesized peptide derived from human ATG16L1, corresponding to a region within C-terminal amino acids.
- Q676U5 A16L1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSSGLRAADFPRWKRHISEQLRRRDRLQRQAFEEIILQYNKLLEKSDLHSVLAQKLQAEKHDVPNRHEISPGHDGTWNDNQLQEMAQLRIKHQEELTELHKKRGELAQLVIDLNNQMQRKDREMQMNEAKIAECLQTISDLETECLDLRTKLCDLERANQTLKDEYDALQITFTALEGKLRKTTEENQELVTRWMAEKAQEANRLNAENEKDSRRRQARLQKELAEAAKEPLPVEQDDDIEVIVDETSDHTEETSPVRAISRAATKRLSQPAGGLLDSITNIFGRRSVSSFPVPQDNVDTHPGSGKEVRVPATALCVFDAHDGEVNAVQFSPGSRLLATGGMDRRVKLWEVFGEKCEFKGSLSGSNAGITSIEFDSAGSYLLAASNDFASRIWTVDDYRLRHTLTGHSGKVLSAKFLLDNARIVSGSHDRTLKLWDLRSKVCIKTVFAGSSCNDIVCTEQCVMSGHFDKKIRFWDIRSESIVREMELLGKITALDLNPERTELLSCSRDDLLKVIDLRTNAIKQTFSAPGFKCGSDWTRVVFSPDGSYVAAGSAEGSLYIWSVLTGKVEKVLSKQHSSSINAVAWSPSGSHVVSVDKGCKAVLWAQY
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Plays an essential role in autophagy: interacts with ATG12-ATG5 to mediate the conjugation of phosphatidylethanolamine (PE) to LC3 (MAP1LC3A, MAP1LC3B or MAP1LC3C), to produce a membrane-bound activated form of LC3 named LC3-II. Thereby, controls the elongation of the nascent autophagosomal membrane. Regulates mitochondrial antiviral signaling (MAVS)-dependent type I interferon (IFN-I) production. Negatively regulates NOD1- and NOD2-driven inflammatory cytokine response. Instead, promotes with NOD2 an autophagy-dependent antibacterial pathway. Plays a role in regulating morphology and function of Paneth cell.
Proteolytic cleavage by activated CASP3 leads to degradation and may regulate autophagy upon cellular stress and apoptotic stimuli.
Phosphorylation at Ser-139 promotes association with the ATG12-ATG5 conjugate to form the ATG12-ATG5-ATG16L1 complex.
Cytoplasm. Preautophagosomal structure membrane>Peripheral membrane protein.
Note: Recruited to omegasomes membranes by WIPI2. Omegasomes are endoplasmic reticulum connected strutures at the origin of preautophagosomal structures. Localized to preautophagosomal structure (PAS) where it is involved in the membrane targeting of ATG5. Localizes also to discrete punctae along the ciliary axoneme.
Belongs to the WD repeat ATG16 family.
Research Fields
· Cellular Processes > Transport and catabolism > Autophagy - other. (View pathway)
· Cellular Processes > Transport and catabolism > Autophagy - animal. (View pathway)
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
References
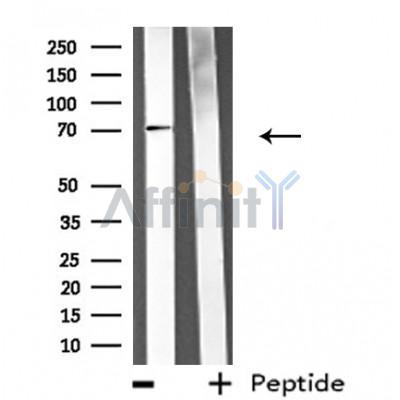
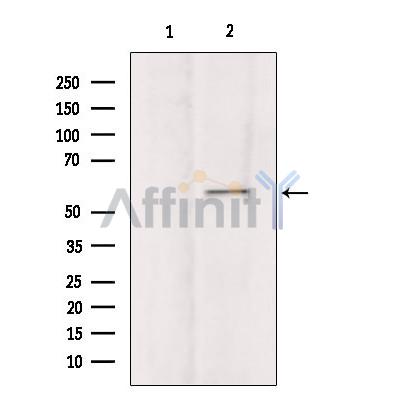
Application: WB Species: Mouse Sample: brain tissue
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.