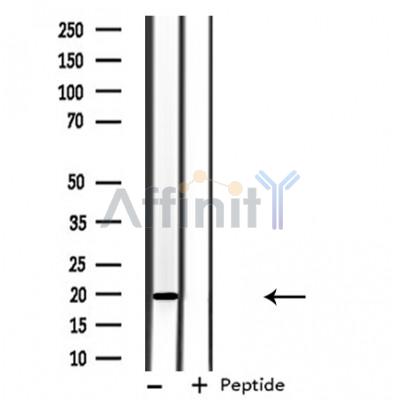
ARF6 Antibody - #DF3746
| Product: | ARF6 Antibody |
| Catalog: | DF3746 |
| Description: | Rabbit polyclonal antibody to ARF6 |
| Application: | WB IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 20 KD; 20kD(Calculated). |
| Uniprot: | P62330 |
| RRID: | AB_2836110 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF3746, RRID:AB_2836110.
Fold/Unfold
antibody;ADP ribosylation factor 6; ADP ribosylation factor protein 6; ADP-ribosylation factor 6; ARF6; ARF6_HUMAN; DKFZp564M0264; Small GTP binding protein; Small GTPase;
Immunogens
A synthesized peptide derived from human ARF6, corresponding to a region within the internal amino acids.
- P62330 ARF6_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGKVLSKIFGNKEMRILMLGLDAAGKTTILYKLKLGQSVTTIPTVGFNVETVTYKNVKFNVWDVGGQDKIRPLWRHYYTGTQGLIFVVDCADRDRIDEARQELHRIINDREMRDAIILIFANKQDLPDAMKPHEIQEKLGLTRIRDRNWYVQPSCATSGDGLYEGLTWLTSNYKS
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
GTP-binding protein involved in protein trafficking that regulates endocytic recycling and cytoskeleton remodeling. Required for normal completion of mitotic cytokinesis (By similarity). Plays a role in the reorganization of the actin cytoskeleton and the formation of stress fibers (By similarity). Involved in the regulation of dendritic spine development, contributing to the regulation of dendritic branching and filopodia extension. Plays an important role in membrane trafficking, during junctional remodeling and epithelial polarization. Regulates surface levels of adherens junction proteins such as CDH1 (By similarity). Required for NTRK1 sorting to the recycling pathway from early endosomes (By similarity).
(Microbial infection) Functions as an allosteric activator of the cholera toxin catalytic subunit, an ADP-ribosyltransferase.
Cytoplasm>Cytosol. Cell membrane>Lipid-anchor. Endosome membrane>Lipid-anchor. Recycling endosome membrane>Lipid-anchor. Cell projection>Filopodium membrane>Lipid-anchor. Cell projection>Ruffle. Cleavage furrow. Midbody>Midbody ring. Early endosome membrane>Lipid-anchor. Golgi apparatus>trans-Golgi network membrane>Lipid-anchor.
Note: Distributed uniformly on the plasma membrane, as well as throughout the cytoplasm during metaphase. Subsequently concentrated at patches in the equatorial region at the onset of cytokinesis, and becomes distributed in the equatorial region concurrent with cleavage furrow ingression. In late stages of cytokinesis, concentrates at the midbody ring/Flemming body (PubMed:23603394). Recruitment to the midbody ring requires both activation by PSD/EFA6A and interaction with KIF23/MKLP1 (By similarity). After abscission of the intercellular bridge, incorporated into one of the daughter cells as a midbody remnant and localizes to punctate structures beneath the plasma membrane (PubMed:23603394). Recruited to the cell membrane in association with CYTH2 and ARL4C. Colocalizes with DAB2IP at the plasma membrane and endocytic vesicles (By similarity). Myristoylation is required for proper localization to membranes (PubMed:7589240).
Ubiquitous, with higher levels in heart, substantia nigra, and kidney.
Belongs to the small GTPase superfamily. Arf family.
Research Fields
· Cellular Processes > Transport and catabolism > Endocytosis. (View pathway)
· Environmental Information Processing > Signal transduction > Ras signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Phospholipase D signaling pathway. (View pathway)
· Organismal Systems > Immune system > Fc gamma R-mediated phagocytosis. (View pathway)
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.