PTTG1 Antibody - #AF0354
| Product: | PTTG1 Antibody |
| Catalog: | AF0354 |
| Description: | Rabbit polyclonal antibody to PTTG1 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IHC |
| Reactivity: | Human, Mouse |
| Prediction: | Pig, Bovine, Sheep, Rabbit, Dog |
| Mol.Wt.: | 30kDa; 22kD(Calculated). |
| Uniprot: | O95997 |
| RRID: | AB_2833520 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0354, RRID:AB_2833520.
Fold/Unfold
AW555095; C87862; Cut2; EAP 1; EAP1; ESP1 associated protein 1; Esp1-associated protein; hPTTG; MGC126883; MGC138276; Pds1; Pituitary tumor transforming 1; Pituitary tumor transforming protein 1; Pituitary tumor-transforming 1, isoform CRA_a; Pituitary tumor-transforming 1, isoform CRA_b; Pituitary tumor-transforming gene 1; Pituitary tumor-transforming gene 1 protein; PTTG 1; PTTG; PTTG1; PTTG1 protein; PTTG1_HUMAN; Pttg3; Securin; Tumor transforming 1; Tumor transforming protein 1; Tumor-transforming protein 1; TUTR 1; TUTR1;
Immunogens
A synthesized peptide derived from human PTTG1, corresponding to a region within the internal amino acids.
Expressed at low level in most tissues, except in adult testis, where it is highly expressed. Overexpressed in many patients suffering from pituitary adenomas, primary epithelial neoplasias, and esophageal cancer.
- O95997 PTTG1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MATLIYVDKENGEPGTRVVAKDGLKLGSGPSIKALDGRSQVSTPRFGKTFDAPPALPKATRKALGTVNRATEKSVKTKGPLKQKQPSFSAKKMTEKTVKAKSSVPASDDAYPEIEKFFPFNPLDFESFDLPEEHQIAHLPLSGVPLMILDEERELEKLFQLGPPSPVKMPSPPWESNLLQSPSSILSTLDVELPPVCCDIDI
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Regulatory protein, which plays a central role in chromosome stability, in the p53/TP53 pathway, and DNA repair. Probably acts by blocking the action of key proteins. During the mitosis, it blocks Separase/ESPL1 function, preventing the proteolysis of the cohesin complex and the subsequent segregation of the chromosomes. At the onset of anaphase, it is ubiquitinated, conducting to its destruction and to the liberation of ESPL1. Its function is however not limited to a blocking activity, since it is required to activate ESPL1. Negatively regulates the transcriptional activity and related apoptosis activity of TP53. The negative regulation of TP53 may explain the strong transforming capability of the protein when it is overexpressed. May also play a role in DNA repair via its interaction with Ku, possibly by connecting DNA damage-response pathways with sister chromatid separation.
Phosphorylated at Ser-165 by CDK1 during mitosis.
Phosphorylated in vitro by ds-DNA kinase.
Ubiquitinated through 'Lys-11' linkage of ubiquitin moieties by the anaphase promoting complex (APC) at the onset of anaphase, conducting to its degradation. 'Lys-11'-linked ubiquitination is mediated by the E2 ligase UBE2C/UBCH10.
Cytoplasm. Nucleus.
Expressed at low level in most tissues, except in adult testis, where it is highly expressed. Overexpressed in many patients suffering from pituitary adenomas, primary epithelial neoplasias, and esophageal cancer.
The N-terminal destruction box (D-box) acts as a recognition signal for degradation via the ubiquitin-proteasome pathway.
The TEK-boxes are required for 'Lys-11'-linked ubiquitination and facilitate the transfer of the first ubiquitin and ubiquitin chain nucleation. TEK-boxes may direct a catalytically competent orientation of the UBE2C/UBCH10-ubiquitin thioester with the acceptor lysine residue.
Belongs to the securin family.
Research Fields
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Cellular Processes > Cell growth and death > Oocyte meiosis. (View pathway)
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
References
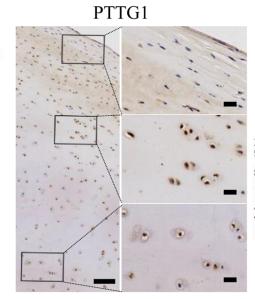
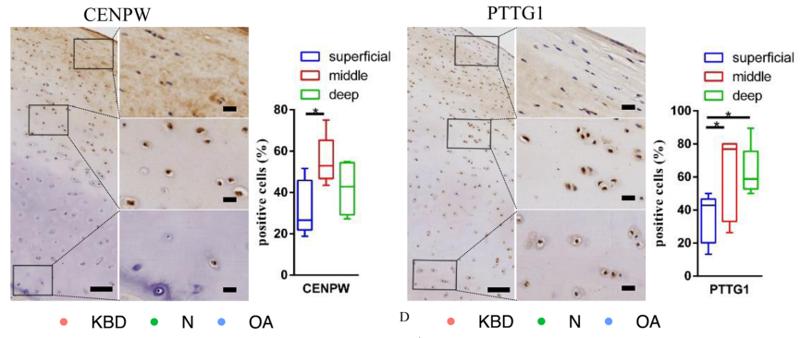
Application: IHC Species: Human Sample: cartilage tissues
Application: IHC Species: human Sample: cartilage
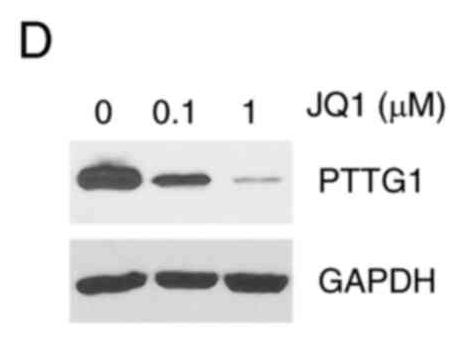
Application: WB Species: Human Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.