YAP Antibody - #DF3182
| Product: | YAP Antibody |
| Catalog: | DF3182 |
| Description: | Rabbit polyclonal antibody to YAP |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Horse, Sheep, Rabbit, Chicken, Xenopus |
| Mol.Wt.: | 65~78kD; 54kD(Calculated). |
| Uniprot: | P46937 |
| RRID: | AB_2835405 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF3182, RRID:AB_2835405.
Fold/Unfold
65 kDa Yes associated protein; 65 kDa Yes-associated protein; COB1; YAp 1; YAP 65; YAP; YAP1; YAP1_HUMAN; YAP2; YAP65; yes -associated protein delta; Yes associated protein 1 65kDa; Yes associated protein 1; Yes associated protein 2; yes associated protein beta; YKI; Yorkie homolog;
Immunogens
A synthesized peptide derived from human YAP, corresponding to a region within the internal amino acids.
Increased expression seen in some liver and prostate cancers. Isoforms lacking the transactivation domain found in striatal neurons of patients with Huntington disease (at protein level).
- P46937 YAP1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MDPGQQPPPQPAPQGQGQPPSQPPQGQGPPSGPGQPAPAATQAAPQAPPAGHQIVHVRGDSETDLEALFNAVMNPKTANVPQTVPMRLRKLPDSFFKPPEPKSHSRQASTDAGTAGALTPQHVRAHSSPASLQLGAVSPGTLTPTGVVSGPAATPTAQHLRQSSFEIPDDVPLPAGWEMAKTSSGQRYFLNHIDQTTTWQDPRKAMLSQMNVTAPTSPPVQQNMMNSASGPLPDGWEQAMTQDGEIYYINHKNKTTSWLDPRLDPRFAMNQRISQSAPVKQPPPLAPQSPQGGVMGGSNSNQQQQMRLQQLQMEKERLRLKQQELLRQAMRNINPSTANSPKCQELALRSQLPTLEQDGGTQNPVSSPGMSQELRTMTTNSSDPFLNSGTYHSRDESTDSGLSMSSYSVPRTPDDFLNSVDEMDTGDTINQSTLPSQQNRFPDYLEAIPGTNVDLGTLEGDGMNIEGEELMPSLQEALSSDILNDMESVLAATKLDKESFLTWL
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Transcriptional regulator which can act both as a coactivator and a corepressor and is the critical downstream regulatory target in the Hippo signaling pathway that plays a pivotal role in organ size control and tumor suppression by restricting proliferation and promoting apoptosis. The core of this pathway is composed of a kinase cascade wherein STK3/MST2 and STK4/MST1, in complex with its regulatory protein SAV1, phosphorylates and activates LATS1/2 in complex with its regulatory protein MOB1, which in turn phosphorylates and inactivates YAP1 oncoprotein and WWTR1/TAZ. Plays a key role in tissue tension and 3D tissue shape by regulating cortical actomyosin network formation. Acts via ARHGAP18, a Rho GTPase activating protein that suppresses F-actin polymerization. Plays a key role to control cell proliferation in response to cell contact. Phosphorylation of YAP1 by LATS1/2 inhibits its translocation into the nucleus to regulate cellular genes important for cell proliferation, cell death, and cell migration. The presence of TEAD transcription factors are required for it to stimulate gene expression, cell growth, anchorage-independent growth, and epithelial mesenchymal transition (EMT) induction.
Isoform 2 and isoform 3 can activate the C-terminal fragment (CTF) of ERBB4 (isoform 3).
Phosphorylated by LATS1 and LATS2; leading to cytoplasmic translocation and inactivation. Phosphorylated by ABL1; leading to YAP1 stabilization, enhanced interaction with TP73 and recruitment onto proapoptotic genes; in response to DNA damage. Phosphorylation at Ser-400 and Ser-403 by CK1 is triggered by previous phosphorylation at Ser-397 by LATS proteins and leads to YAP1 ubiquitination by SCF(beta-TRCP) E3 ubiquitin ligase and subsequent degradation. Phosphorylated at Thr-119, Ser-138, Thr-154, Ser-367 and Thr-412 by MAPK8/JNK1 and MAPK9/JNK2, which is required for the regulation of apoptosis by YAP1.
Ubiquitinated by SCF(beta-TRCP) E3 ubiquitin ligase.
Cytoplasm. Nucleus.
Note: Both phosphorylation and cell density can regulate its subcellular localization. Phosphorylation sequesters it in the cytoplasm by inhibiting its translocation into the nucleus. At low density, predominantly nuclear and is translocated to the cytoplasm at high density (PubMed:18158288, PubMed:20048001). PTPN14 induces translocation from the nucleus to the cytoplasm (PubMed:22525271).
Increased expression seen in some liver and prostate cancers. Isoforms lacking the transactivation domain found in striatal neurons of patients with Huntington disease (at protein level).
The first coiled-coil region mediates most of the interaction with TEAD transcription factors.
Belongs to the YAP1 family.
Research Fields
· Environmental Information Processing > Signal transduction > Hippo signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hippo signaling pathway - multiple species. (View pathway)
References
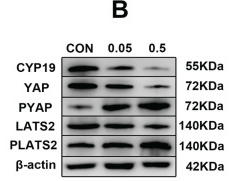
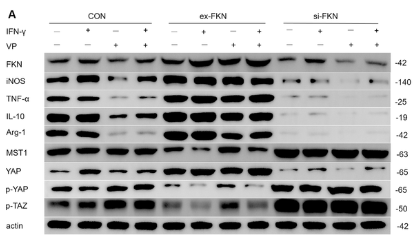
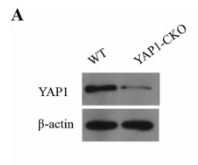
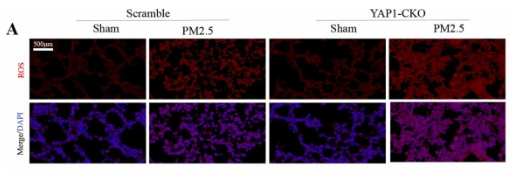
Application: WB Species: Rat Sample: H9C2 cells
Application: WB Species: Mouse Sample:
Application: IF/ICC Species: Mouse Sample: lung tissues
Application: WB Species: Rat Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.