ATM Antibody - #AF4119
| Product: | ATM Antibody |
| Catalog: | AF4119 |
| Description: | Rabbit polyclonal antibody to ATM |
| Application: | WB IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat, Monkey |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog |
| Mol.Wt.: | 351kDa; 351kD(Calculated). |
| Uniprot: | Q13315 |
| RRID: | AB_2835361 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF4119, RRID:AB_2835361.
Fold/Unfold
A-T mutated; A-T mutated homolog; AT mutated; AT1; ATA; Ataxia telangiectasia mutated; Ataxia telangiectasia mutated gene; Ataxia telangiectasia mutated homolog (human); Ataxia telangiectasia mutated homolog; ATC; ATD; ATDC; ATE; ATM; ATM serine/threonine kinase; ATM_HUMAN; DKFZp781A0353; MGC74674; OTTHUMP00000232981; Serine protein kinase ATM; Serine-protein kinase ATM; Serine/threonine-protein kinase ATM; Tefu; TEL1; TEL1, telomere maintenance 1, homolog; TELO1; Telomere fusion protein;
Immunogens
A synthesized peptide derived from human ATM, corresponding to a region within N-terminal amino acids.
Found in pancreas, kidney, skeletal muscle, liver, lung, placenta, brain, heart, spleen, thymus, testis, ovary, small intestine, colon and leukocytes.
- Q13315 ATM_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSLVLNDLLICCRQLEHDRATERKKEVEKFKRLIRDPETIKHLDRHSDSKQGKYLNWDAVFRFLQKYIQKETECLRIAKPNVSASTQASRQKKMQEISSLVKYFIKCANRRAPRLKCQELLNYIMDTVKDSSNGAIYGADCSNILLKDILSVRKYWCEISQQQWLELFSVYFRLYLKPSQDVHRVLVARIIHAVTKGCCSQTDGLNSKFLDFFSKAIQCARQEKSSSGLNHILAALTIFLKTLAVNFRIRVCELGDEILPTLLYIWTQHRLNDSLKEVIIELFQLQIYIHHPKGAKTQEKGAYESTKWRSILYNLYDLLVNEISHIGSRGKYSSGFRNIAVKENLIELMADICHQVFNEDTRSLEISQSYTTTQRESSDYSVPCKRKKIELGWEVIKDHLQKSQNDFDLVPWLQIATQLISKYPASLPNCELSPLLMILSQLLPQQRHGERTPYVLRCLTEVALCQDKRSNLESSQKSDLLKLWNKIWCITFRGISSEQIQAENFGLLGAIIQGSLVEVDREFWKLFTGSACRPSCPAVCCLTLALTTSIVPGTVKMGIEQNMCEVNRSFSLKESIMKWLLFYQLEGDLENSTEVPPILHSNFPHLVLEKILVSLTMKNCKAAMNFFQSVPECEHHQKDKEELSFSEVEELFLQTTFDKMDFLTIVRECGIEKHQSSIGFSVHQNLKESLDRCLLGLSEQLLNNYSSEITNSETLVRCSRLLVGVLGCYCYMGVIAEEEAYKSELFQKAKSLMQCAGESITLFKNKTNEEFRIGSLRNMMQLCTRCLSNCTKKSPNKIASGFFLRLLTSKLMNDIADICKSLASFIKKPFDRGEVESMEDDTNGNLMEVEDQSSMNLFNDYPDSSVSDANEPGESQSTIGAINPLAEEYLSKQDLLFLDMLKFLCLCVTTAQTNTVSFRAADIRRKLLMLIDSSTLEPTKSLHLHMYLMLLKELPGEEYPLPMEDVLELLKPLSNVCSLYRRDQDVCKTILNHVLHVVKNLGQSNMDSENTRDAQGQFLTVIGAFWHLTKERKYIFSVRMALVNCLKTLLEADPYSKWAILNVMGKDFPVNEVFTQFLADNHHQVRMLAAESINRLFQDTKGDSSRLLKALPLKLQQTAFENAYLKAQEGMREMSHSAENPETLDEIYNRKSVLLTLIAVVLSCSPICEKQALFALCKSVKENGLEPHLVKKVLEKVSETFGYRRLEDFMASHLDYLVLEWLNLQDTEYNLSSFPFILLNYTNIEDFYRSCYKVLIPHLVIRSHFDEVKSIANQIQEDWKSLLTDCFPKILVNILPYFAYEGTRDSGMAQQRETATKVYDMLKSENLLGKQIDHLFISNLPEIVVELLMTLHEPANSSASQSTDLCDFSGDLDPAPNPPHFPSHVIKATFAYISNCHKTKLKSILEILSKSPDSYQKILLAICEQAAETNNVYKKHRILKIYHLFVSLLLKDIKSGLGGAWAFVLRDVIYTLIHYINQRPSCIMDVSLRSFSLCCDLLSQVCQTAVTYCKDALENHLHVIVGTLIPLVYEQVEVQKQVLDLLKYLVIDNKDNENLYITIKLLDPFPDHVVFKDLRITQQKIKYSRGPFSLLEEINHFLSVSVYDALPLTRLEGLKDLRRQLELHKDQMVDIMRASQDNPQDGIMVKLVVNLLQLSKMAINHTGEKEVLEAVGSCLGEVGPIDFSTIAIQHSKDASYTKALKLFEDKELQWTFIMLTYLNNTLVEDCVKVRSAAVTCLKNILATKTGHSFWEIYKMTTDPMLAYLQPFRTSRKKFLEVPRFDKENPFEGLDDINLWIPLSENHDIWIKTLTCAFLDSGGTKCEILQLLKPMCEVKTDFCQTVLPYLIHDILLQDTNESWRNLLSTHVQGFFTSCLRHFSQTSRSTTPANLDSESEHFFRCCLDKKSQRTMLAVVDYMRRQKRPSSGTIFNDAFWLDLNYLEVAKVAQSCAAHFTALLYAEIYADKKSMDDQEKRSLAFEEGSQSTTISSLSEKSKEETGISLQDLLLEIYRSIGEPDSLYGCGGGKMLQPITRLRTYEHEAMWGKALVTYDLETAIPSSTRQAGIIQALQNLGLCHILSVYLKGLDYENKDWCPELEELHYQAAWRNMQWDHCTSVSKEVEGTSYHESLYNALQSLRDREFSTFYESLKYARVKEVEEMCKRSLESVYSLYPTLSRLQAIGELESIGELFSRSVTHRQLSEVYIKWQKHSQLLKDSDFSFQEPIMALRTVILEILMEKEMDNSQRECIKDILTKHLVELSILARTFKNTQLPERAIFQIKQYNSVSCGVSEWQLEEAQVFWAKKEQSLALSILKQMIKKLDASCAANNPSLKLTYTECLRVCGNWLAETCLENPAVIMQTYLEKAVEVAGNYDGESSDELRNGKMKAFLSLARFSDTQYQRIENYMKSSEFENKQALLKRAKEEVGLLREHKIQTNRYTVKVQRELELDELALRALKEDRKRFLCKAVENYINCLLSGEEHDMWVFRLCSLWLENSGVSEVNGMMKRDGMKIPTYKFLPLMYQLAARMGTKMMGGLGFHEVLNNLISRISMDHPHHTLFIILALANANRDEFLTKPEVARRSRITKNVPKQSSQLDEDRTEAANRIICTIRSRRPQMVRSVEALCDAYIILANLDATQWKTQRKGINIPADQPITKLKNLEDVVVPTMEIKVDHTGEYGNLVTIQSFKAEFRLAGGVNLPKIIDCVGSDGKERRQLVKGRDDLRQDAVMQQVFQMCNTLLQRNTETRKRKLTICTYKVVPLSQRSGVLEWCTGTVPIGEFLVNNEDGAHKRYRPNDFSAFQCQKKMMEVQKKSFEEKYEVFMDVCQNFQPVFRYFCMEKFLDPAIWFEKRLAYTRSVATSSIVGYILGLGDRHVQNILINEQSAELVHIDLGVAFEQGKILPTPETVPFRLTRDIVDGMGITGVEGVFRRCCEKTMEVMRNSQETLLTIVEVLLYDPLFDWTMNPLKALYLQQRPEDETELHPTLNADDQECKRNLSDIDQSFNKVAERVLMRLQEKLKGVEEGTVLSVGGQVNLLIQQAIDPKNLSRLFPGWKAWV
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Serine/threonine protein kinase which activates checkpoint signaling upon double strand breaks (DSBs), apoptosis and genotoxic stresses such as ionizing ultraviolet A light (UVA), thereby acting as a DNA damage sensor. Recognizes the substrate consensus sequence [ST]-Q. Phosphorylates 'Ser-139' of histone variant H2AX at double strand breaks (DSBs), thereby regulating DNA damage response mechanism. Also plays a role in pre-B cell allelic exclusion, a process leading to expression of a single immunoglobulin heavy chain allele to enforce clonality and monospecific recognition by the B-cell antigen receptor (BCR) expressed on individual B-lymphocytes. After the introduction of DNA breaks by the RAG complex on one immunoglobulin allele, acts by mediating a repositioning of the second allele to pericentromeric heterochromatin, preventing accessibility to the RAG complex and recombination of the second allele. Also involved in signal transduction and cell cycle control. May function as a tumor suppressor. Necessary for activation of ABL1 and SAPK. Phosphorylates DYRK2, CHEK2, p53/TP53, FANCD2, NFKBIA, BRCA1, CTIP, nibrin (NBN), TERF1, RAD9, UBQLN4 and DCLRE1C. May play a role in vesicle and/or protein transport. Could play a role in T-cell development, gonad and neurological function. Plays a role in replication-dependent histone mRNA degradation. Binds DNA ends. Phosphorylation of DYRK2 in nucleus in response to genotoxic stress prevents its MDM2-mediated ubiquitination and subsequent proteasome degradation. Phosphorylates ATF2 which stimulates its function in DNA damage response. Phosphorylates ERCC6 which is essential for its chromatin remodeling activity at DNA double-strand breaks.
Phosphorylated by NUAK1/ARK5. Autophosphorylation on Ser-367, Ser-1893, Ser-1981 correlates with DNA damage-mediated activation of the kinase. During the late stages of DNA damage response, dephosphorylated following deacetylation by SIRT7, leading to ATM deactivation.
Acetylation, on DNA damage, is required for activation of the kinase activity, dimer-monomer transition, and subsequent autophosphorylation on Ser-1981. Acetylated in vitro by KAT5/TIP60. Deacetylated by SIRT7 during the late stages of DNA damage response, promoting ATM dephosphorylation and subsequent deactivation.
Nucleus. Cytoplasmic vesicle.
Note: Primarily nuclear. Found also in endocytic vesicles in association with beta-adaptin.
Found in pancreas, kidney, skeletal muscle, liver, lung, placenta, brain, heart, spleen, thymus, testis, ovary, small intestine, colon and leukocytes.
The FATC domain is required for interaction with KAT5.
Belongs to the PI3/PI4-kinase family. ATM subfamily.
Research Fields
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Cellular Processes > Cell growth and death > p53 signaling pathway. (View pathway)
· Cellular Processes > Cell growth and death > Apoptosis. (View pathway)
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Environmental Information Processing > Signal transduction > NF-kappa B signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Genetic Information Processing > Replication and repair > Homologous recombination.
· Human Diseases > Drug resistance: Antineoplastic > Platinum drug resistance.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
References
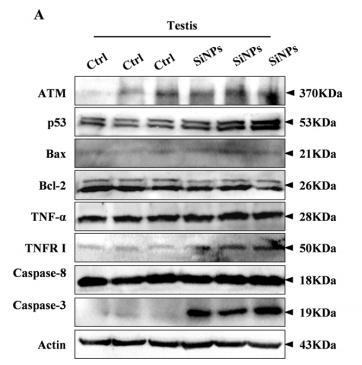
Application: WB Species: mouse Sample: testis
Application: WB Species: Mouse Sample: GC-1 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.